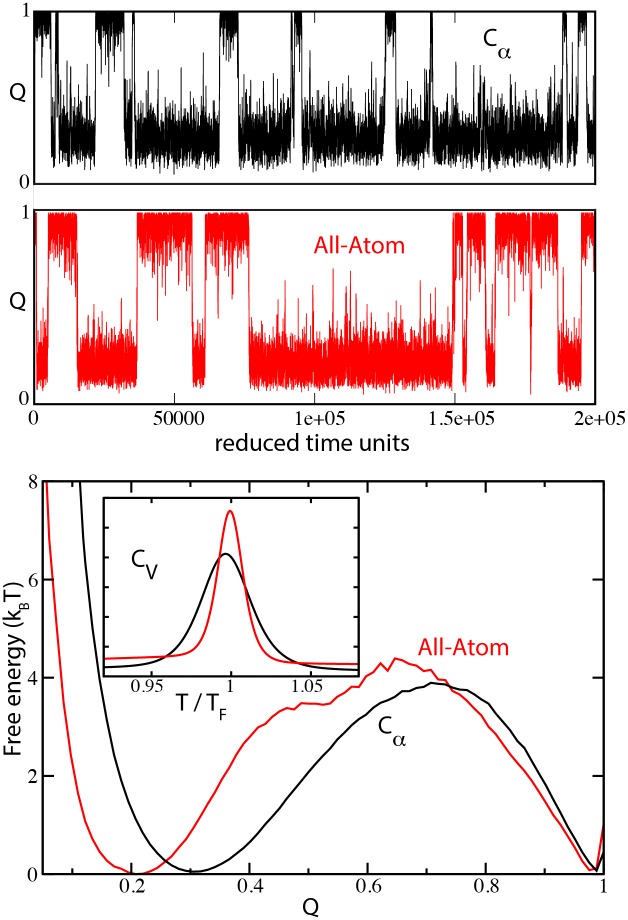
Fig 1. Protein folding simulations with the default Cα and all-atom models of the 64 residue chymotrypsin inhibitor 2 (PDB code: 1YPA).
Top: Folding trajectories near folding temperature (TF) of the Cα (black) and all-atom (red) models. Bottom: Free energy as a function of , the number of native Cα pairs within 1.5 times their native distance. The same coordinate is used to describe both models. Inset: Specific heat for the two models (normalized to have equal area). TF in reduced units for the all-atom model is 0.97 and for the Cα model is 1.17 (117 and 140 in the GROMACS .mdp file, respectively).