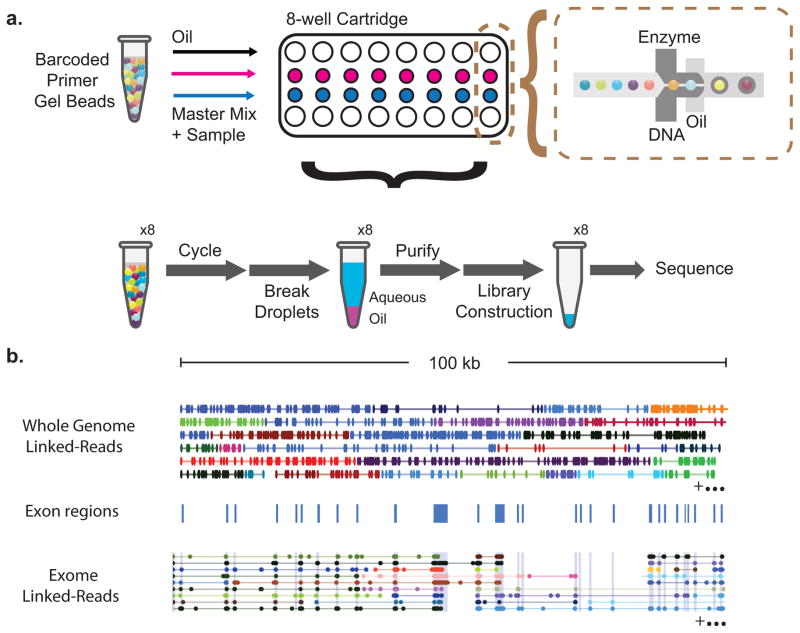
Figure 1. Overview of the technology for generating linked-reads.
(a) Gel beads loaded with primers and barcoded oligonucleotides are first mixed with DNA and enzyme mixture, and subsequently mixed with oil-surfactant solution at a microfluidic “double-cross” junction. Gel bead-containing droplets flow to a reservoir where gel beads are dissolved, initiating whole genome primer extension. The products are pooled from each droplet. The final library preparation requires shearing the libraries and incorporation of Illumina adapters. (b) Top panel, linked-reads of the ALK gene from the NA12878 WGS sample. Each line represents linked-reads with the same barcode, with dots representing reads, and color depicting reads with different barcodes. Middle panel, blue blocks showing exon boundaries of the ALK gene. Bottom panel, linked-reads of ALK gene from the NA12878 exome data. Although there are only reads in exon regions, reads from neighboring exons are linked because of common barcodes. Only a very small fraction of linked-reads is presented here to conserve space.