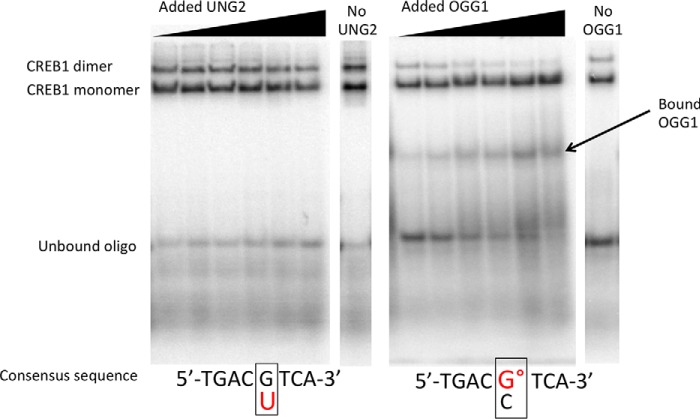
FIGURE 1.
Monomer and dimer CREB1 binding to the CRE site contained within a ds 39-mer oligonucleotide are cleanly resolved by EMSA and are differentially impacted by UNG and OGG1. In these representative gel images, UNG2 and CREB1, or OGG1 and CREB1, were mixed together followed by the addition of the labeled oligonucleotide (oligo) and incubated for 90 min at 4 °C prior to EMSA analysis. Note detectable OGG1 binding to the °G-containing oligonucleotide. Apparent reduction in unbound substrate at low concentrations of OGG1 is due to direct binding by the repair glycosylase. On the other hand, binding of UNG2 to U-containing CRE site is not sufficiently stable to detect in the absence of CREB1. Typical gels were obtained when UNG2 was added to substrate containing a G/U mispair (left) or when OGG1 was added to substrate containing a °G/C pair (right) in the CpG site of CRE.