FIGURE 1.
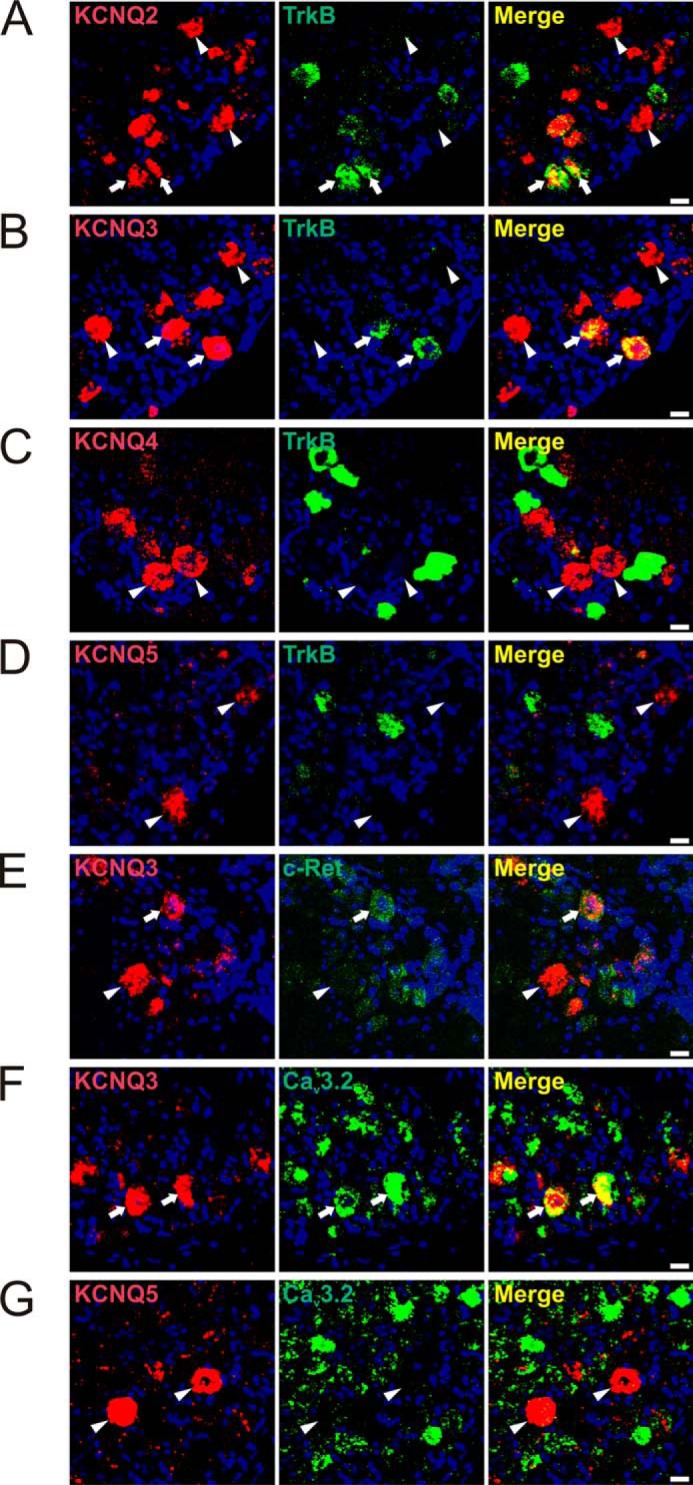
KCNQ2 and -3, but not KCNQ4 and -5, are expressed in a subset of TrkB-positive, low-threshold D-hair fibers. Expression of different KCNQ channels in DRG sections was revealed by double in situ hybridization. A and B, co-expression of KCNQ2 and KCNQ3 with TrkB, respectively (arrows). A, 27 out of 102 KCNQ2-positive and of 53 TrkB-positive cells were co-labeled for both proteins. B, out of 190 KCNQ3-positive neurons and 84 TrkB-positive cells, 71 were labeled for both TrkB and KCNQ3. C, KCNQ4-expressing neurons were not labeled for TrkB (n = 89). D, TrkB-labeling is also absent in KCNQ5-positive cells (n = 71). E, 26 out of 72 KCNQ3-positive cells were labeled for c-Ret (arrows). F, co-expression of KCNQ3 and Cav3.2. 35 out of 71 KCNQ3-positive neurons were labeled for Cav3.2 (arrows). G, 114 out of 131 KCNQ5-positive neurons were not labeled for Cav3.2. Arrowheads, KCNQ2-, -3-, -4-, and -5-positive DRG neurons, which were not labeled for TrkB (A–D), c-Ret (E), or Cav3.2 (G), respectively. Nuclei were labeled with DAPI (blue). Scale bars: 20 μm.
