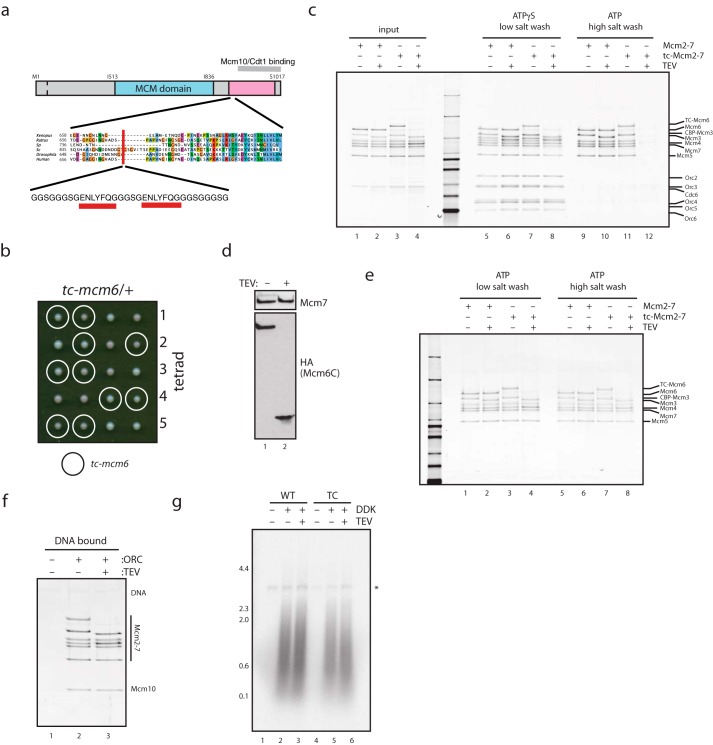
FIGURE 3.
The C terminus of Mcm6 is not required for Mcm10 recruitment, or DNA replication initiation. a, schematic of the tc-Mcm6 construct developed in this study. b, diploid budding yeast cells containing one copy each of wild-type and TEV-cleavable MCM6 were sporulated, and tetrads were analyzed for growth, indicating that TEV cleavage site insertion has no effect on viability. c, purified MCM·Cdt1 or tc-MCM·Cdt1 heptamers (which contain tc-Mcm6) were incubated with TEV protease and used in double hexamer assembly assays in the presence of ATP or the poorly hydrolyzed ATP analogue ATPγS. DNA-bound proteins were analyzed by SDS-PAGE and silver stain. Although all complexes were recruited to DNA under low salt conditions in the presence ATPγS (lanes 5–8), TEV cleavage of tc-Mcm6 prevented high salt (0.5 m NaCl)-resistant MCM loading (lane 12). Note that CBP-Mcm3 shifts in mobility ±TEV protease due to a TEV cleavage site between CBP and Mcm3. d, Mcm2–7 containing tc-Mcm6 was loaded onto DNA with purified proteins and incubated with TEV protease as indicated. DNA-bound proteins were separated by SDS-PAGE and analyzed by Western blotting. e, MCM and tc-MCM were loaded onto DNA, washed with high salt buffer, cut with TEV protease as indicated, and washed with low salt (0.3 m potassium glutamate) or high salt buffer as indicated. f, MCM and tc-MCM were loaded onto DNA, washed with high salt, cut with TEV protease as indicated, washed with low salt, and mixed with purified Mcm10. DNA was washed, and bound proteins were analyzed by SDS-PAGE and silver stain. g, MCM and tc-MCM were loaded onto DNA, cut with TEV protease as indicated, and used in a DNA replication system reconstituted with purified proteins (13). Radiolabeled replication products were separated by denaturing alkaline agarose electrophoresis and visualized by autoradiography. The asterisk marks nicked plasmid products that are labeled in a replication-independent manner.