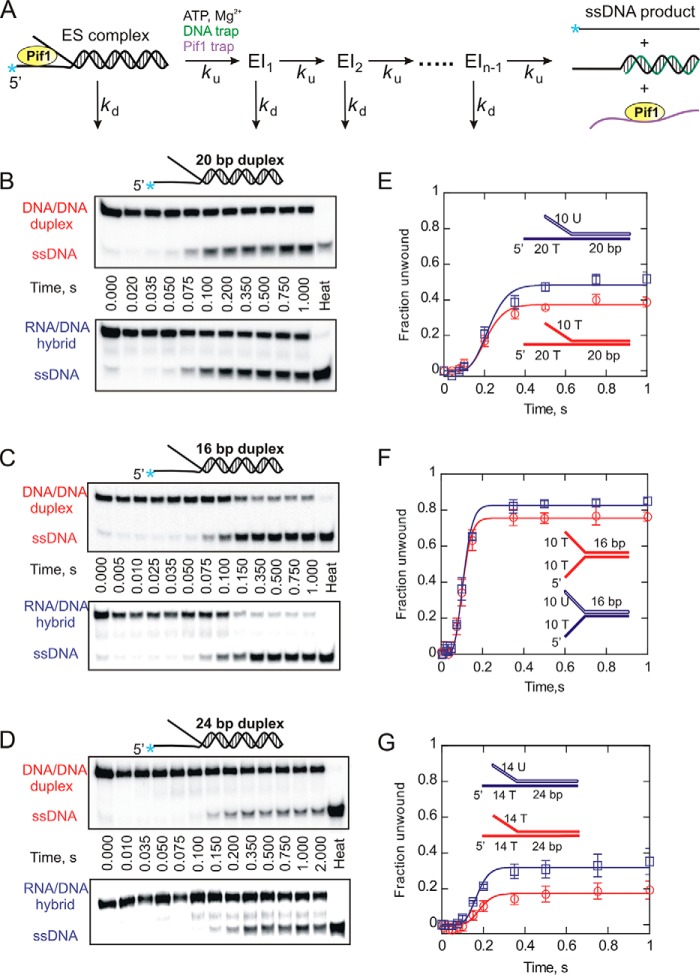
FIGURE 1.
Helicase-catalyzed unwinding kinetics for DNA:DNA (red circles) and RNA:DNA (blue squares) forked duplexes under single turnover (single cycle) conditions. A, a schematic representation of a single cycle unwinding reaction. Pif1 takes a series of n sequential steps to separate duplex strands. ES, enzyme-substrate complex; EI, intermediate. Each step is defined by an unwinding rate constant (ku) and a dissociation rate constant (kd). Unwinding reactions were performed under excess enzyme conditions as described under “Experimental Procedures.” B–D, the representative 20% native PAGE gels for 20- (B), 16- (C), and 24-bp (D) forked substrates show the separation of ssDNA product from forked duplexes. Product formation was plotted against time using Kaleidagraph software. E–G, shown are the reaction progress curves for 20- (E), 16- (F), and 24-bp (G) forked substrates. The data were fit to the scheme shown in A using Kintek Explorer (29). Error bars indicate the standard deviation of 3 independent experiments. Kinetic constants are shown in Tables 2 and 3.