FIGURE 5.
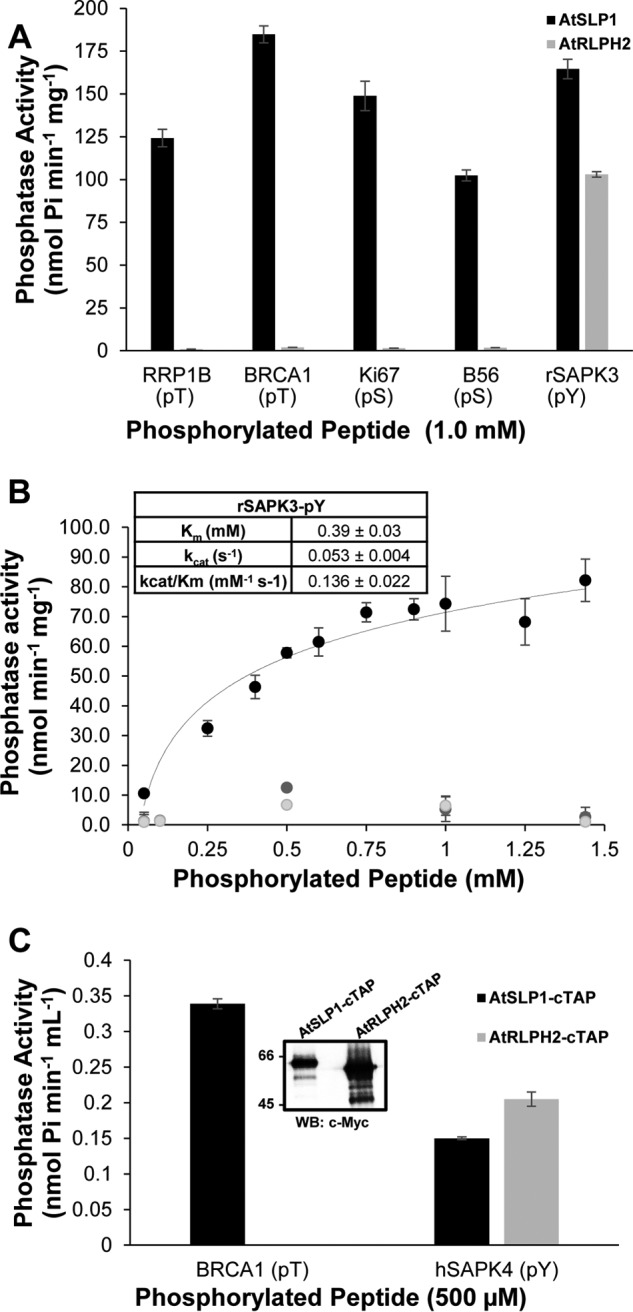
Phospho-peptide substrate preferences of purified A. thaliana bacteria-like PPP family protein phosphatases. A. thaliana SLP1 (AtSLP1, black bars) and RLPH2 (AtRLPH2, gray bars) were expressed and purified as tagged fusion proteins in E. coli. A, 1 μg of each purified phosphatase was incubated with 1 mm designated phosphorylated peptide at 30 °C for 1 h to determine their phospho-substrate preference. The phospho-residue is indicated below the peptide name. pT, pThr; pS, pSer; pY, pTyr. B, bacterially expressed tagged AtRLPH2 kinetic constants were determined by performing assays with increasing amounts of rSAPK3 peptide (pTyr; black), or rSAPK3 peptide with a pSer (light gray) or pThr (dark gray) substituted for the pTyr. Only the pTyr peptide yielded adequate activity to calculate Km, kcat, and kcat/Km values, which are presented in the inset. C, constitutively expressed AtSLP1-cTAP (black bars) and AtRLPH2-cTAP (gray bars) were isolated from A. thaliana rosette leaf tissue as described under “Experimental Procedures.” Incubation of each TAP purified protein with either 500 μm BRCA1 (pThr) or 500 μm hSAPK4 (pTyr) phospho-peptides revealed each protein phosphatase substrate preference. The inset shows a Western blot (WB) of the Myc epitope of the TAP tag on each protein to demonstrate that equal amounts of phosphatase were used in each assay. Phosphatase activity was determined using a stop time malachite green assay. Error bars represent S.E. (n = 3).
