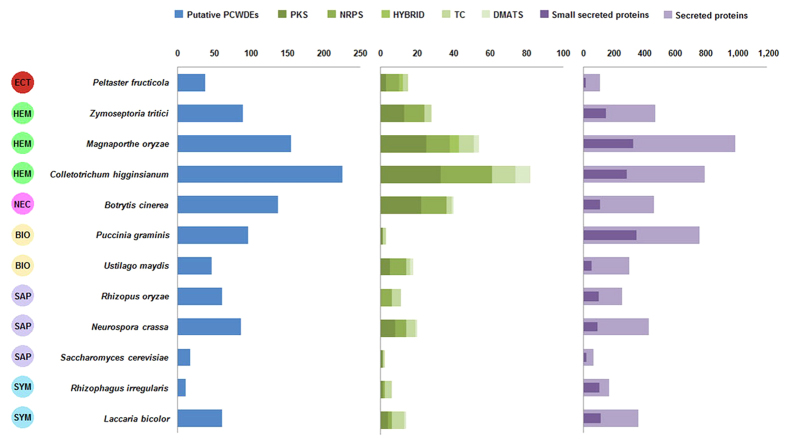
Figure 2. The numbers of genes encoding putative plant cell wall-degrading enzymes, key secondary metabolite synthetases and secreted proteins identified in the genomes of P. fructicola and 11 additional fungal species included in this study.
The boxes on the left represent the life style of the selected organisms. ECT, ectophyte; HEM, hemibiotrophs; NEC, necrotroph; BIO, biotrophs; SAP, saprotrophs; SYM, symbionts. The colored bars representing the secondary metabolic enzymes are identified by the key at the top. PKS, polyketide synthases; NRPS, nonribosomal peptide synthases; TC, terpene cyclase; DMATS, dimethyl allyl tryptophan synthases; HYBRID, PKS-NRPS hybrids.