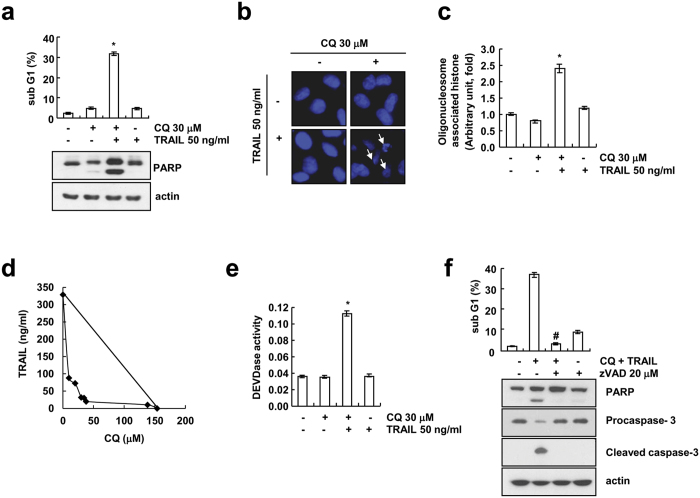
Figure 1. CQ sensitizes TRAIL-mediated apoptosis in human renal cancer Caki cells.
(a–c) Caki cells were treated with the 50 ng/ml TRAIL in the presence or absence of 30 μM chloroquine (CQ) for 24 h. Apoptosis was analyzed as the sub-G1 fraction by FACS analysis. The protein expression of PARP was determined by Western blotting. The level of actin was used as a loading control (a). The condensation and fragmentation of the nuclei were detected by 4′,6′-diamidino-2-phenylindole (DAPI) staining (b). DNA fragmentation was determined using a DNA fragmentation detection kit (c). (d) Isoboles were obtained by plotting the combined concentrations of each drug required to produce 50% cell death. The straight line connecting the IC50 values obtained for two agents when applied alone corresponds to an additivity of their independent effects. Values below this line indicate synergy, whereas values above this line indicate antagonism. (e) Caki cells were treated with the 50 ng/ml TRAIL in the presence or absence of 30 μM chloroquine (CQ) for 24 h. Enzymatic activities of DEVDase were determined by incubation of 20 μg of total protein with 200 μM chromogenic substrate (DEVD-pNA) in a 100 μl assay buffer for 2 h at 37 °C. The release of chromophore p-nitroanilide (pNA) was monitored spectrophotometrically (405 nm). (f) Caki cells were pre-treated with 20 μM z-VAD-fmk (zVAD) for 30 min before combined treatment with 30 μM CQ and 50 ng/ml TRAIL for 24 h. Apoptosis was analyzed as the sub-G1 fraction by FACS analysis. The protein expression of PARP, procaspase-3 and cleaved caspase-3 were determined by Western blotting. The level of actin was used as a loading control. The values in panel (a,c,e,f) represent the mean ± SD from three independent samples. *p < 0.01 compared to the control. #p < 0.01 compared to the CQ plus TRAIL.