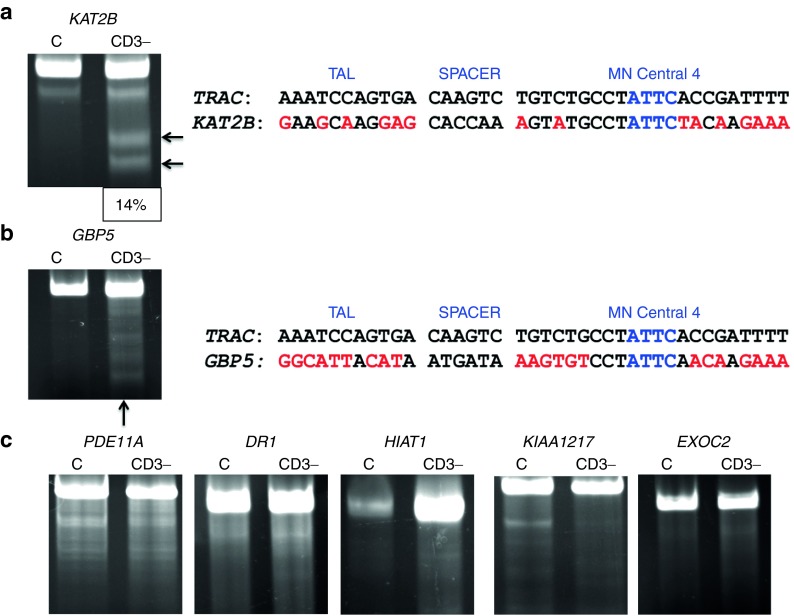
Figure 8.
Off-target assessment in primary T-cells. Target loci were amplified from 100% CD3-null cells and analyzed by the Surveyor assay for evidence of nuclease activity. (a) KAT2B Surveyor cleavage products and sequence alignment to TRAC. Cleavage products are indicated by arrows and quantitative gel analysis indicating is shown at bottom of gel. (b) GBP5 Surveyor image and genomic sequence in relation to TRAC. (c) PDE11A, DR1, HIAT1, KIAA1217, and EXOC2 Surveyor analysis that did not reveal MT OT cleavage. DR1, downregulator of transcription 1; EXOC2, exocyst complex component 2, TBP-binding (negative cofactor); GBP5, guanylate-binding protein 5; HIAT1, hippocampus abundant transcript 1; KAT2B, K(Lysine) acetyltransferase 2B; KIAA1217, sickle tail; PDE11A, phosphodiesterase 11A. Black arrows indicate cleavage products following Surveyor/CELII enzyme digest. The sequence alignments are in relation to the TAL repeat variable diresidues, the intervening spacer sequence separating the TAL from the meganuclease domain, and the meganuclease domain with the I-OnuI homing endonuclease “central four” ATTC sequence. Mismatches between the TRAC and OT sites are lighter shaded letters.