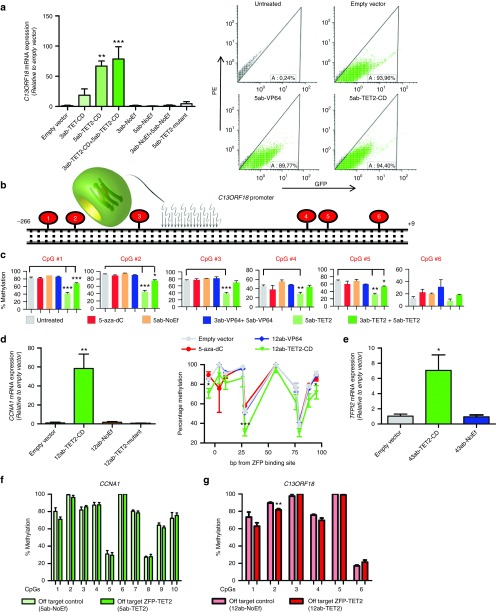
Figure 5.
TET2-mediated gene activation and demethylation. (a) Relative C13ORF18 mRNA expression (left) after retrovirally induced expression of ZFPs carrying GFP and either TET2-CD, TET2-mutant or NoEf. On the right, the GFP positivity of transduced cells as exemplified for untreated cells or cells expressing an empty vector, 5ab-VP64 or 5ab-TET2 measured by FACS. (b) Schematic representation of the C13ORF18 promoter region carrying the target site of 5ab-TET2-CD and six CpGs surrounding this site. (c) The DNA methylation status of the six CpGs in the C13ORF18 promoter after expressing the gene-targeting constructs or treatment with 5-aza-dC (5 µM for 3 days (n = 3 samples from Figure 1d–f)) as quantified by bisulfite pyrosequencing. Values represent the mean of at least two independent experiments ± SEM. (d) Relative CCNA1 expression after retrovirally induced expression of ZFPs carrying either TET2-CD, TET2-mutant or NoEf in CaSki cells (left). On the right, the DNA methylation status of 10 CpGs in the CCNA1 promoter after expressing the gene-targeting constructs 12ab-TET2-CD and 12ab-VP64 (or treatment with 5-aza-dC (5 µM for 3 days (n = 3)) as quantified by bisulfite pyrosequencing. Statistical differences were determined between empty vector and 12ab-TET2-CD. (e) Relative TFPI2 mRNA expression after retrovirally induced expression of ZFPs carrying either TET2-CD or NoEf in HeLa cells. All cells were GFP sorted (except NoEf) after transduction and re-transduced before analysis. Expression of C13ORF18, CCNA1, and TFPI2 is normalized to GAPDH and relative to an empty vector. mRNA was quantified by qPCR and each bar represents the mean of at least three independent experiments measured in triplicate ± SEM. (f) The off-target effect of the C13ORF18-targeting 5ab-TET2 ZFP and the NoEf control on the methylation status of the CCNA1 promoter. (g) The off-target effect of the CCNA1-targeting 12ab-TET2 ZFP and the NoEf control on the methylation status of the C13ORF18 promoter. Methylation levels were quantified by bisulfite pyrosequencing and each data point represents the mean of three independent experiments ± SEM