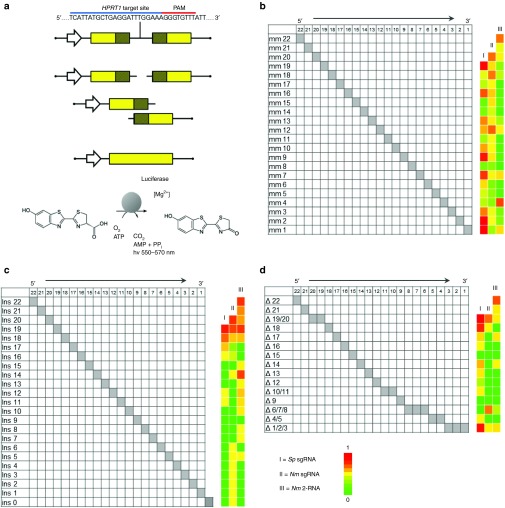
Figure 2.
Comparison of mismatch tolerance between Nme and Spy CRISPR-Cas9 systems. (a) Schematic of the luciferase based single-strand annealing (SSA) assay used to determine gRNA specificity. Following successful cleavage of the target site by Cas9, the shaded homologous regions can recombine via SSA resulting in the restoration of the full length luciferase open reading frame and expression of luciferase. The level of luciferase expressed is measured by the amount of D-luciferin which is converted into oxyluciferin and light. The activity of gRNA variants containing mismatches (b), insertions (c), or deletions (d) are displayed as heatmaps based on relative single-strand annealing activity. The activity of all gRNA variants was normalized to the original guide sequence. gRNAs of 20, 21, and 23 nt were used for Spy sgRNA, Nme sgRNA, and Nme 2-RNA systems respectively. All data derived from biological triplicates. The sequences and raw data for all gRNA variants can be found in Supplementary Table S3.