Figure 1.
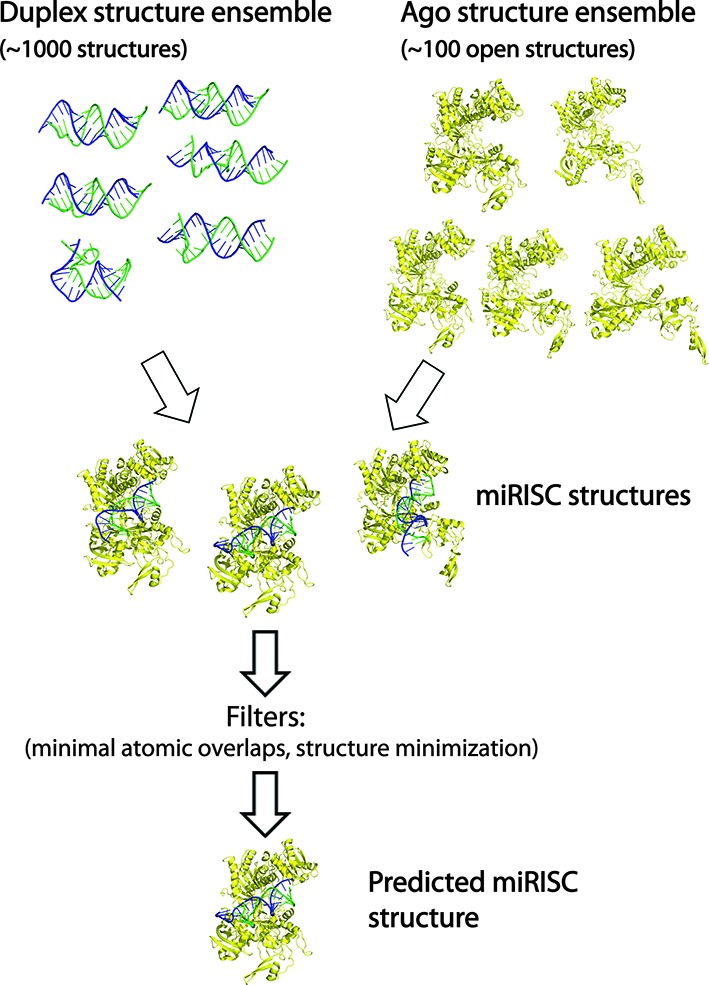
An algorithm for assembling miRISC structures. First, duplex and Ago structure ensembles are independently generated using MC–Sym duplex assembly and ENM methods, respectively. Second, each duplex structure is docked (i.e. guide RNA strand alignment using 3HJF as a reference complex) to each Ago conformation to produce an ensemble of model miRISC structures. Third, the miRISC structures are ranked by the number of atomic overlaps and then the top scoring complexes are energy minimized, which are the predicted structures.
