Figure 5.
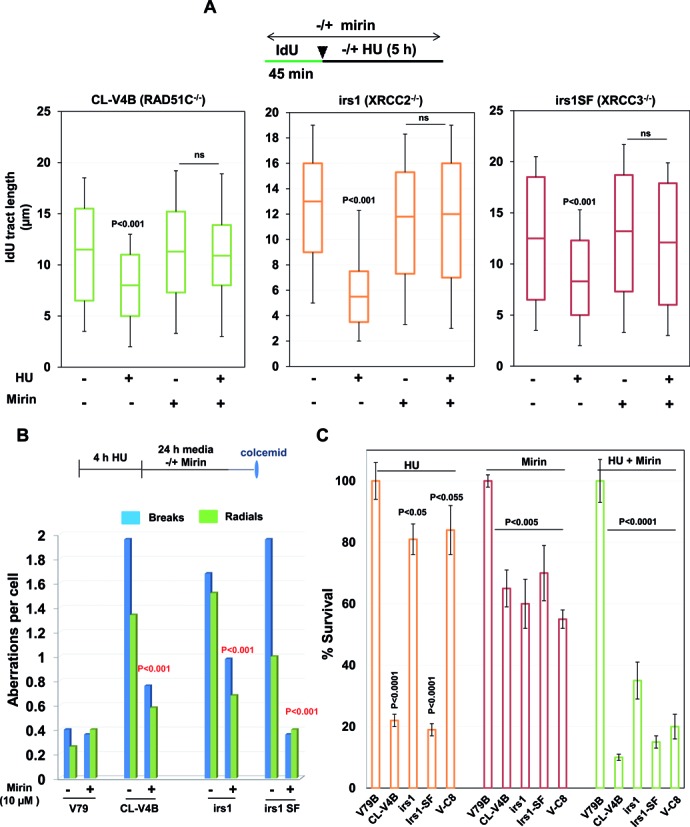
RAD51 paralogs suppress MRE11-mediated extensive fork processing of stalled replication forks. (A) Experimental design for analysis of replication fork stability in RAD51 paralogs-deficient cells after chemical inhibition of MRE11 nuclease. IdU labeled DNA fibers were measured in the indicated cells following mock or HU treatment with or without inhibition of MRE11 with Mirin. Bar graphs represent median IdU tract length from 200 analyzed DNA fibers. (B) Quantification of chromosomal aberrations in the form of chromatid breaks and radial chromosomes in RAD51 paralog deficient cells with or without inhibition of MRE11. (C) Cellular sensitivity of RAD51C, XRCC2, XRCC3 and BRCA2 deficient cells to HU treatment following MRE11 inhibition. Indicated cells were treated with or without 10 μM Mirin, followed by 2 mM HU for 4 h and cell survival was determined. Results shown are mean ± SD of three independent experiments. P-values are obtained for paralog deficient cells in comparison with parental cells. (CL-V4B- RAD51C−/−, irs1- XRCC2−/−, irs1SF- XRCC3−/−, V-C8- BRCA2−/− and V79B, V79, and CHO-AA8 are parental cell lines for CL-V4B, irs1 and irs1SF respectively).
