Figure 8.
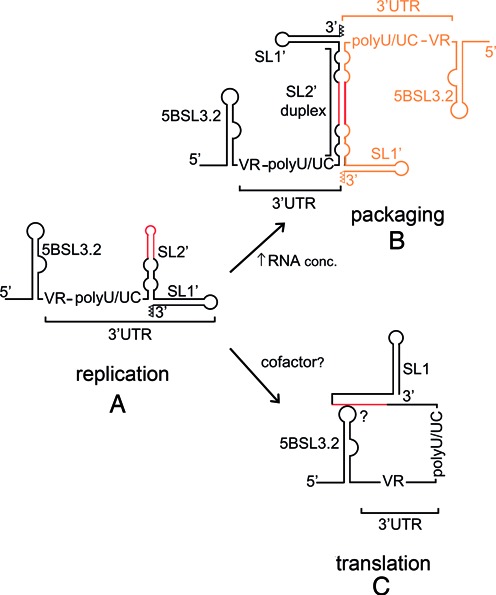
Scheme summarizing the structural features of domain 3′X supported by this study, and their possible impact on the HCV life cycle. (A) Preferred structure of the domain monomers, comprising stem–loops SL2’ and SL1’. This structure contains unpaired nt (represented as open triangles) at the 3′ terminus of stem SL1’ and promotes viral RNA replication. (B) With sufficient RNA concentration domain 3′X forms homodimers comprising an extended 110-nt SL2’ duplex flanked by intramolecular SL1’ hairpins. These structures probably mediate self-association and packaging of viral genomes. (C) The 3′X RNA can interact with an upstream 5BSL3.2 stem–loop. This contact, reported to favor translation, involves local melting of the upper stem of SL2’ and may be aided by protein cofactors. In (A), (B) and (C), the DLS tract is colored red; in (B), the second HCV genome is represented in orange.
