Figure 6.
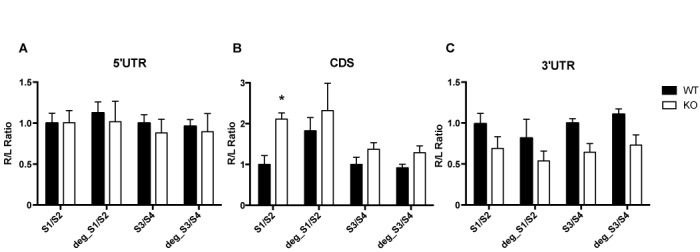
Influence of FMRP on luciferase reporter gene assay bearing G4RIF. psiCHECK-2 plasmid expressing Renilla bearing S1/S2 or S3/S4 or DegS1/S2 or DegS3/S4 G4RIF subregions were transfected in STEK cells. Three independent experiments with three replicates for each transfection, were quantified. For each transfection Renilla luciferase activity (R) was normalized with Firefly luciferase activity (L). In order to facilitate the comparison between native and degenerated sequences in WT and KO cells, ratios for all conditions were divided by the mean of the signal obtained for the cognate native sequences in the WT cells. Results are presented as the mean ±SEM (two way ANOVA, *P < 0.05). (A) S1/S2 or S3/S4 or DegS1/S2 or DegS3/S4 sequences have been cloned in the 5′ UTR of the Renilla reporter; (B) S1/S2 or S3/S4 or DegS1/S2 or DegS3/S4 sequences are cloned in frame with the coding sequence of the reporter gene; (C) S1/S2 or S3/S4 or DegS1/S2 or DegS3/S4 sequences are cloned in the 3′ UTR of the reporter gene.
