Figure 7.
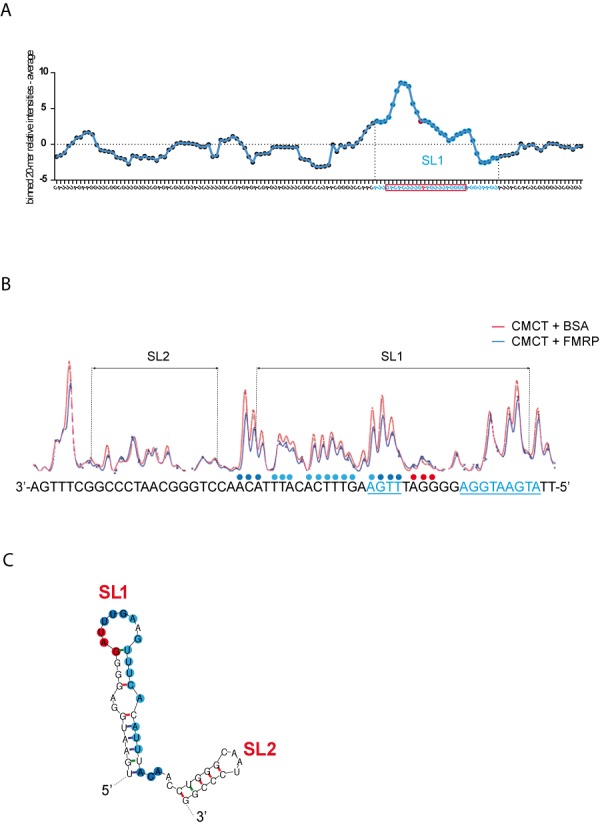
FMRP protection of SL1. (A) Relative CMCT reactivities along the G4RIF sequence. We used the QuShape software to quantify the reactivities of each nucleotide toward the CMCT probe in the presence of BSA or FMRP. BSA reactivities were subtracted to those in the presence of FMRP, providing a relative reactivity of each nucleotide to the CMCT probe in the presence of FMRP along the whole G4RIF sequence (Supplementary Table SI). We then summed the relative reactivities for 20 consecutive nucleosides to identify the sequence that is protected by FMRP. The relative reactivities of these 20-mer bins were plotted as a function of the G4RIF sequence. A sharp increase in the computed relative reactivities, highlighting the protection by FMRP, corresponds to the SL1 part of G4RIF sequence that is represented with blue dots on the graph. An exemple of a 20mer bin, centered on a « A » whose relative reactivity is shown as a red plot on the graph, is represented by the red rectangle at the bottom of the figure. (B) CMCT reactivities along the SL1 and SL2 part of the G4RIF in the presence of BSA (red line) or FMRP (blue line) are displayed with the corresponding sequence at the bottom of the figure. In the presence of FMRP (1 μM), a 20 nt sequence at the 3' end of SL1 is less reactive toward the CMCT probe than in the presence of BSA (1 μM). Colored dots above the sequence represent the relative protection by FMRP. Dark blue dots represent a stronger protection than light blue dots. Red dots represent a higher sensitivity to the CMCT probe in the presence of FMRP. ACUK/WGGA sequences are displayed in blue and underlined. (C) RNA secondary structure model of SL1/SL2 sequence showing results from chemical modification experiments in the presence of FMRP. Dark blue dots represent a stronger protection than light blue dots. Red dots represent a higher sensitivity to CMCT probe in the presence of FMRP.
