Figure 2.
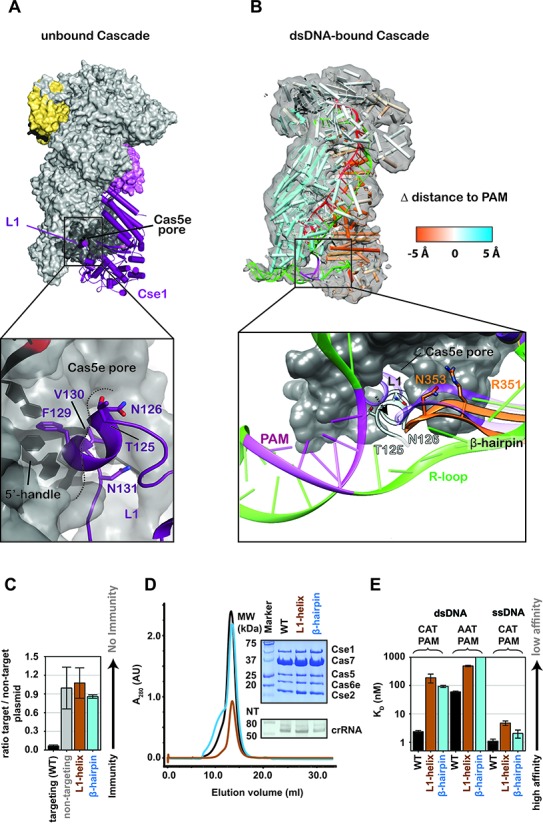
The L1-helix and a long β-hairpin in the Cse1 subunit are involved in protospacer adjacent motif (PAM) recognition and duplex destabilization. (A) In the structure of Cascade prior to DNA binding, L1 sits inside the Cas5e pore. F129, V130 and N131 make contact with nucleobases in the 5′-handle of the crRNA. T125 and N126 are solvent accessible. (B) Molecular Dynamic Flexible Fitting was used to model atomic coordinates of Cascade prior to target binding, into the cryo-EM density of Cascade bound to a dsDNA target with a PAM. The model is colored according to changes in distance relative to the PAM. Motion toward the PAM is colored orange and motion away from the PAM is blue. In the simulation, the L1-helix is positioned proximal to the PAM and the β-hairpin is positioned between single-strand regions of the DNA target. (C) Mutations in the L1-helix (T125N/N126K) or the β-hairpin (R351G/N353P/A355S/S356R) result in immunodeficiency. (D) Elution profile of the L1-helix and β-hairpin mutants. The insert shows a Commassie blue-stained SDS-PAGE gel (top) and a denaturing polyacrylamide gel of phenol-extracted crRNA isolated from each of the Cascade complexes (bottom). (E) Equilibrium dissociation constants of Cascade, the L1-helix and β-hairpin mutants for dsDNA substrates containing either 5′-CAT-3′ or 5′-AAT-3′ PAM and for ssDNA substrates containing a 5′-CAT-3′ PAM.
