Figure 4.
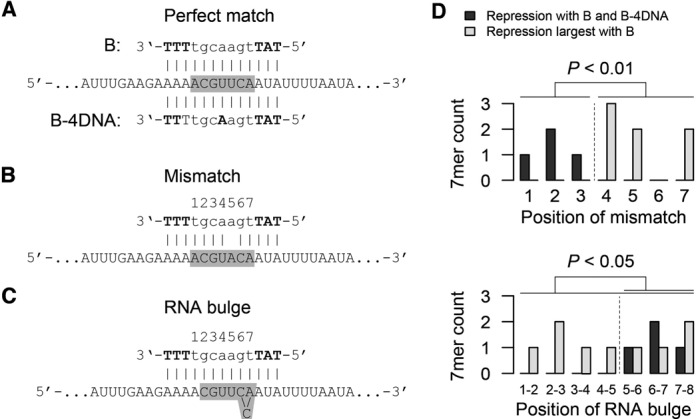
Effective RNA-oligonucleotide duplexes. (A) Perfect match binding of the B and B-4DNA oligonucleotide to the LibSeq RNA sequence. LNA nucleotides are indicated in bold capital letters and the gray box shows the location of the degenerate sequence in the LibSeq library. (B) Specific example of a duplex between the B oligonucleotide and the LibSeq RNA, which affects expression despite a mismatch in position 5. Positions are indicated above the oligonucleotide, LNA nucleotides are indicated in bold capital letters and the gray box shows the location of the degenerate sequence in the LibSeq library. (C) Specific example of a duplex between the B oligonucleotide and the LibSeq RNA, which affects expression despite an RNA bulge between positions 6 and 7. Positions are indicated above the oligonucleotide, LNA nucleotides are indicated in bold capital letters and the gray box shows the location of the degererate sequence in the LibSeq library. (D) Positions of mismatches (top) and RNA bulges (bottom) in LibSeq RNA–oligonucleotide duplexes, which repress expression both for the B and the B-4DNA oligonucleotide (black bars) or only for the B oligonucleotide (gray bars). Both for mismatches and and RNA bulges the positions are significantly different for the two oligonucleotides.
