Figure 1.
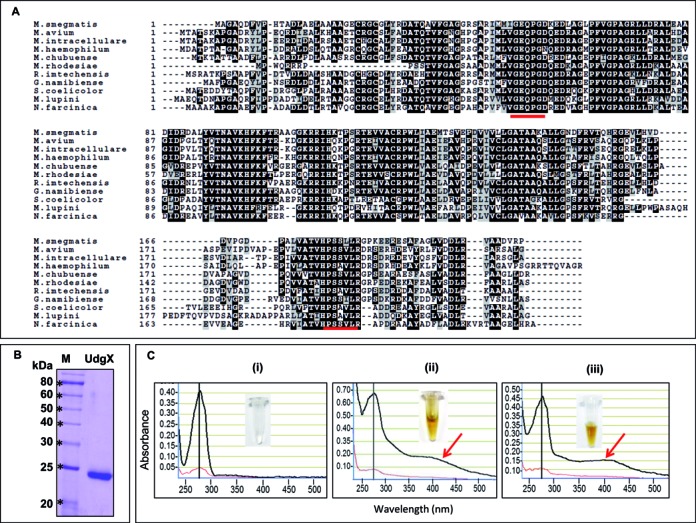
Multiple sequence alignment of MsmUdgX with its homologs, purification of MsmUdgX and its UV-VIS scan. (A) Multiple sequence alignment of MsmUdgX and its homologs from other bacteria using Clustal W and Boxshade server. Identical residues are shown in black whereas the similar residues are shown in light gray boxes. The motif A and motif B sequences have been underlined. NCBI reference sequence ID are: M. smegmatis, WP_011726794.1; M. avium, ZP_05218036.1; M. intracellulare, ZP_05227544.1; M. haemophilum, ZP_21917190.1; M. chubuense, YP_006453151.1; M. rhodesiae, WP_014212855.1; R. imtechensis, ZP_10004147.1; G. namibiense, ZP_10960189.1; S. coelicolor, NP_628659.1; M. lupini, ZP_21028631.1; and N. farcinica, YP_118940.1. (B) 12% SDS-PAGE analysis of MsmUdgX showing its purity. (C) UV-VIS scan of, (i) MsmUng (ii) MsmUdgX, (iii) MtuUdgB. The black and red lines are absorbances at 1 mm and 0.1 mm, respectively. The presence of additional absorbance peaks at 415 nm (arrow) in MsmUdgX and MtuUdgB are indicative of Fe-S cluster. Insets show the colors of the protein stocks.
