Figure 4.
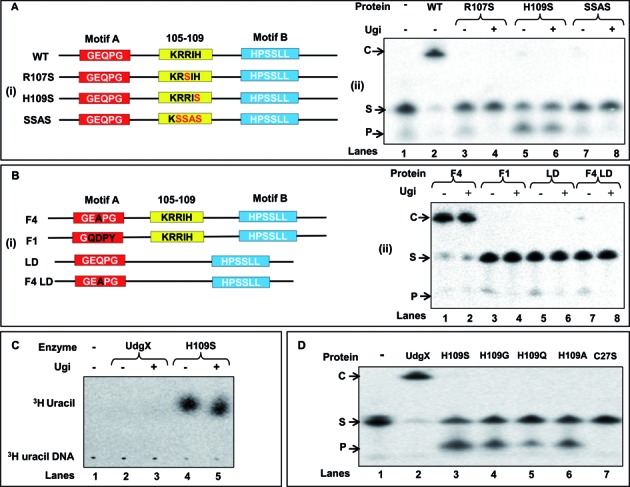
Site directed mutagenesis of the KRRIH region of MsmUdgX and its activity assay. (A (i) and B (i)) Line diagrams of MsmUdgX and its mutants highlighting the regions targeted by site directed mutagenesis. Motif A, motif B and the KRRIH loop region are shown in red, blue and yellow boxes, respectively. (A (ii)) Uracil excision/binding assays with MsmUdgX (WT) and its R107S, H109S and SSAS mutant derivatives. ∼200 ng of each protein was reacted with 5′ 32P-labeled SSU9, the reaction mix was resolved on 8 M urea-PAGE (15%) and analyzed by phosphor imaging. The presence of uracil excision activity is seen by the fast migrating product band, ‘P’ whereas the uracil-DNA binding activity by the slow migrating complex, ‘C’. (B (ii)) Uracil excision/binding assays with MsmUdgX (WT) and its F4, F1, LD and F4 LD mutant derivatives (∼200 ng each) using SSU9. (C) Uracil release assay of MsmUdgX H109S (∼200 ng) using 3H-uracil containing DNA (21 000 c.p.m) as substrate. Released uracil was separated by TLC (CEL 300 PEI/UV254) and seen as a fast migrating species upon phosphor imaging (the radioactive spot comigrated with the cold uracil spot detected by UV shadowing). (D) Uracil excision/binding assay with MsmUdgX and its H109S, H109G, H109Q, H109A and C27S derivatives. ∼200 ng of each protein was used in a standard UDG assay with SSU9 substrate.
