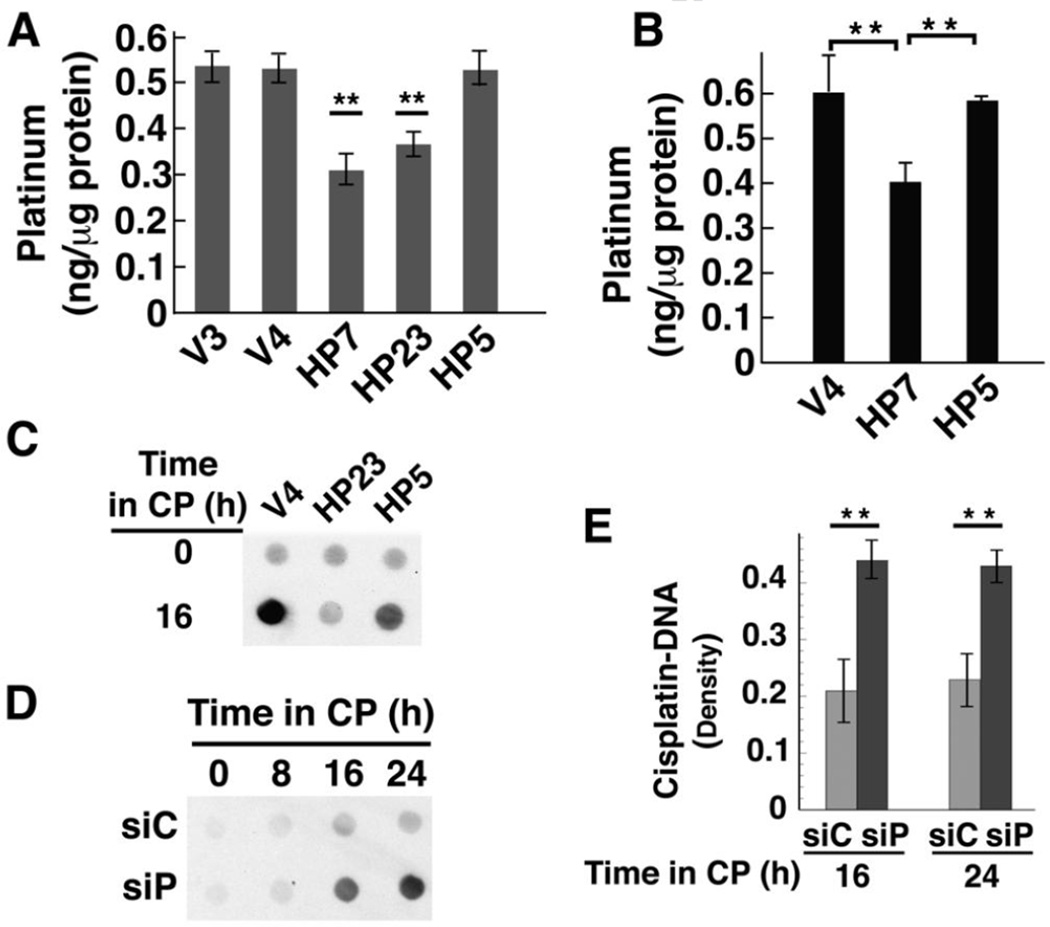
Fig. 6. Cisplatin accumulation and DNA-bound platinum are lower in cells over-expressing active HClpP and higher in cells with reduced HClpP.
A, Cellular accumulation of cisplatin. Cultures were treated with 3.5 µg/ml cisplatin for 16 h, after which the cells were washed and platinum levels in total cell lysates were measured in triplicate by atomic absorption. Error bars represent the SEM in two independent experiments. Platinum levels in HP7 and HP23 cells were between 50–60% of control V3 or V4 levels in two separate experiments (P < 0.01; noted with asterisks). Levels in HP5 were not significantly different from those in control cells (P >0.1). B, Mitochondrial accumulation of cisplatin. Cells were treated with 2.5 µg/ml cisplatin for 16 h, collected by centrifugation and washed. Mitochondria were isolated by differential centrifugation, washed, and dissolved in nitric acid. Platinum in the mitochondrial fraction was quantitated by ICP mass spectrometry. Error bars represent the SEM of the values obtained in two independent experiments. Levels in HP7 cells were 65 ± 2 of those in V4 cells (P < 0.01). C, V4, HP23, and HP5 cells were treated with 3.5 µg/ml cisplatin for 16 h. Total cellular DNA was isolated and 10 µg-aliquots of DNA were spotted and probed for platinum modified DNA using a monoclonal antibody that recognizes diguanosine-cisplatin-adducts. D and E, Accumulation of platinum in total cellular DNA after knockdown of HClpP. Parallel cultures of HeLa cells were transfected with 10 nM HClpP siRNA (siP) or control siRNA (siC) for 5 h, washed to remove the siRNA, and then incubated in DMEM medium for 16 h. Cisplatin (CP) (2.5 µg/ml) was added and individual cultures were harvested at different times. Total DNA was extracted from the cells and purified. D, Equal aliquots (12 µg) from each sample were spotted on a nitrocellulose membrane, and cisplatin adducts were detected as above. Spots contain < 0.1 ng MtDNA, and the signal reflects primarily modification of nuclear DNA. E, Serial dilutions of each DNA sample (12, 6, and 3 µg) were spotted and cisplatin adducts were detected immunochemically. The blots were scanned, and the densities for each sample were normalized for loading and averaged. The error bars represent the SEM of the normalized densities from the diluted samples.