Abstract
Germline stem cell differentiation in Caenorhabditis elegans is controlled by glp-1 Notch signaling. Cell fate regulator GLD-1 is sufficient to induce meiotic entry and expressed at a high level during meiotic prophase, inhibiting mitotic gene activity. glp-1 signaling and other regulators control GLD-1 levels post-transcriptionally (low in stem cells, high in meiotic prophase), but many aspects of GLD-1 regulation are uncharacterized, including the link between glp-1-mediated transcriptional control and post-transcriptional GLD-1 regulation. We established a sensitive assay to quantify GLD-1 levels across an ∼35-cell diameter field, where distal germline stem cells differentiate proximally into meiotic prophase cells in the adult C. elegans hermaphrodite, and applied the approach to mutants in known or proposed GLD-1 regulators. In wild-type GLD-1 levels elevated ∼20-fold in a sigmoidal pattern. We found that two direct transcriptional targets of glp-1 signaling, lst-1 and sygl-1, were individually required for repression of GLD-1. We determined that lst-1 and sygl-1 act in the same genetic pathway as known GLD-1 translational repressor fbf-1, while lst-1 also acts in parallel to fbf-1, linking glp-1-mediated transcriptional control and post-transcriptional GLD-1 repression. Additionally, we estimated the position in wild-type gonads where germ cells irreversibly commit to meiotic development based on GLD-1 levels in worms where glp-1 activity was manipulated to cause an irreversible fate switch. Analysis of known repressors and activators, as well as modeling the sigmoidal accumulation pattern, indicated that regulation of GLD-1 levels is largely regional, which we integrated with the current view of germline stem cell differentiation.
Keywords: germline stem cells, GLP-1 Notch, GLD-1, Caenorhabditis elegans, meiotic entry, post-transcriptional control, cell fate regulation
NOTCH signaling is a highly conserved pathway that contributes to stem cell maintenance and differentiation in a variety of developmental and organismal contexts (Bray 2006; Liu et al. 2010; Andersson et al. 2011). Notch signaling control of stem cell maintenance and differentiation is typified by a source cell, providing ligand to a limited number of recipient, receptor-expressing cells. The Caenorhabditis elegans germline provides a unique context for Notch-mediated control of a stem cell population, where the glp-1 Notch signaling receptor gives rise to the polarized pattern of germline stem cell differentiation. Under optimal growth conditions, germline stem cell differentiation into meiotic prophase spans an ∼30-cell diameter region of >250 cells in the distal region of the adult hermaphrodite gonad (Kimble and Crittenden 2007; Byrd and Kimble 2009; Hansen and Schedl 2013) (Figure 1A). A relatively large pool of stem cells (∼60–80 cells) is maintained through glp-1 signaling activation triggered by ligands expressed by a large and complex somatic gonad cell called the distal tip cell (DTC) (Kimble and White 1981; Austin and Kimble 1987; Henderson et al. 1994; Tax et al. 1994; Nadarajan et al. 2009; Byrd et al. 2014; Fox and Schedl 2015). As germ cells are displaced out of reach of the DTC, glp-1 signaling is thought to drop below a threshold level of activity; then, after completing their ongoing mitotic cell cycle (terminal mitosis), daughters enter meiotic S and subsequently overtly adopt the meiotic fate by beginning leptotene/zygotene (Figure 1A). This polarized stem cell differentiation pattern is ideal for rapid generation of large numbers of meiotic prophase cells under optimal conditions for progeny production (Fox and Schedl 2015).
Figure 1.
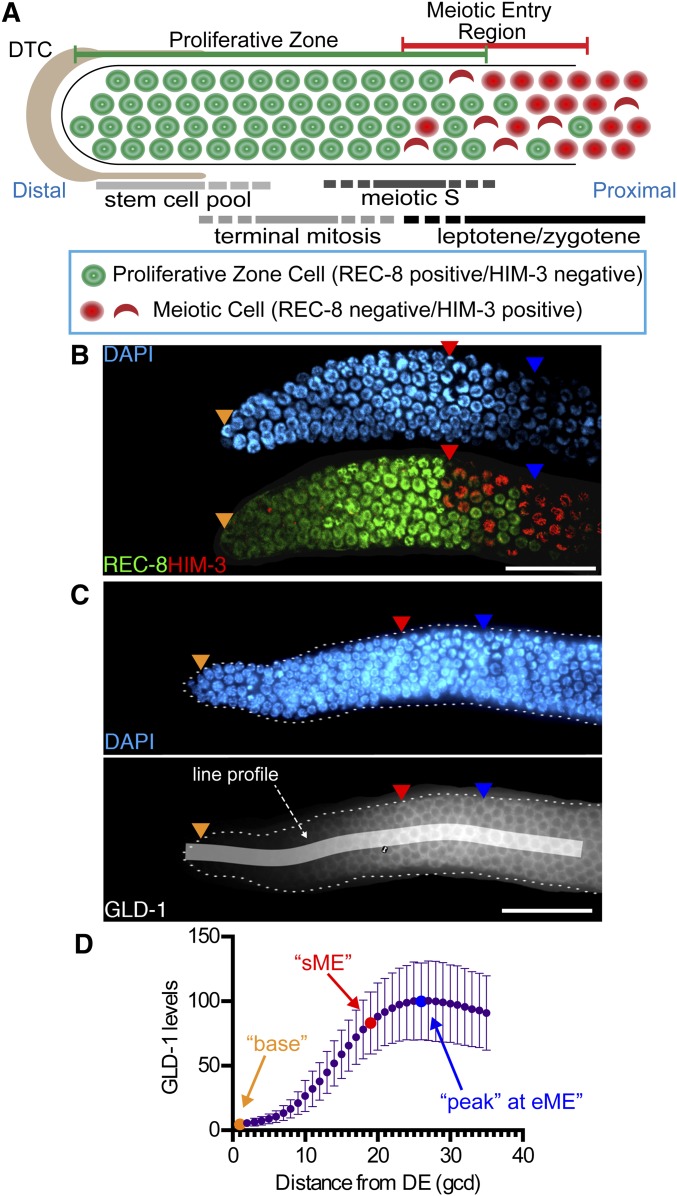
GLD-1 accumulation in the proliferative zone. (A) Schematic of the distal germline from the adult hermaphrodite. The distal proliferative zone, capped by the large somatic distal tip cell (DTC), is ∼20 germ cell diameters (gcd) in length and contains ∼230 germ cells. The proliferative zone is composed of three partially overlapping pools of cells, as indicated. At the proximal end of the proliferative zone, germ cells begin overt meiotic entry. Staining for markers define proliferative zone cells (e.g., nucleoplasmic REC-8 staining) and meiotic prophase cells (e.g., chromosomal HIM-3 staining). First gcd with HIM-3 positive nucleus is start of meiotic entry region (sME) and last REC-8 positive nucleus is end of meiotic entry region (eME). (B) DAPI and REC-8/HIM-3 immunofluorescence image for wild type. (C) Representative DAPI and GLD-1 immunofluorescence image for wild type. Quantified 35-gcd region is depicted as white shaded rectangle. Orange triangles, 1st gcd; red triangles, sME; blue triangles, eME; and white bars are 25 µm. (D) Plot of wild-type GLD-1 levels. DE, distal end. Error bars ± SD. n = 90. Orange dot, base; red dot, sME; blue dot, peak.
The mechanism whereby glp-1 mediates control over a large population of germline stem cells is not well understood. Previous genetic analysis by others and us demonstrated that glp-1 maintains the germline stem cell fate through repression of at least three redundant genetic pathways called the gld-1, gld-2, and third meiotic entry pathways (Kadyk and Kimble 1998; Eckmann et al. 2004; Hansen et al. 2004a; Fox et al. 2011). The gld-1 pathway includes the gld-1 and nos-3 genes. gld-1 encodes an RNA binding protein that inhibits germline stem cell fate and/or promotes meiotic entry through translational repression of mitotic genes (Biedermann et al. 2009; Fox et al. 2011; Jungkamp et al. 2011). nos-3 encodes an RNA binding protein related to Drosophila Nanos (Kraemer et al. 1999), a known translational repressor. nos-3 promotes meiotic entry at least in part through controlling GLD-1 levels and/or activity (Hansen et al. 2004b), but how it mediates this activity is unknown. The gld-2 pathway includes gld-2 and gld-3 (Kadyk and Kimble 1998; Eckmann et al. 2004; Hansen et al. 2004a; Schmid et al. 2009). gld-2 encodes a cytoplasmic poly-A polymerase that promotes translation of meiotic entry genes to inhibit germline stem cell fate and/or promote meiotic entry (Wang et al. 2002; Suh et al. 2006; Kim et al. 2010). gld-3 encodes an RNA binding protein that promotes meiotic entry by facilitating gld-2 interaction with its direct targets (Suh et al. 2006; Schmid et al. 2009). Genetic analysis of mutants lacking both gld-1 and gld-2 pathway genes revealed the existence of at least a third meiotic entry pathway (Hansen et al. 2004a; Fox et al. 2011), but the identity of genes that act in this pathway are currently unknown. Meiotic entry occurs normally in mutants lacking genes representing any one pathway (i.e., gld-1 or gld-2 single mutants), but meiotic entry is impaired in mutants lacking genes from separate pathways (i.e., gld-1gld-2 double mutants), highlighting that these genes are not individually required for meiotic entry.
The activity and or levels of the gld-1 and gld-2 pathway genes are responsive to glp-1 signaling activity. For example, GLD-1 levels are high in distal germ cells in the absence of glp-1, and GLD-1 levels remain low when glp-1 signaling is ectopically high (Hansen et al. 2004b). However, none of the aforementioned genes of the gld-1 and gld-2 meiotic entry pathways are obvious direct transcriptional targets of glp-1 signaling and are thus indirectly repressed by glp-1 signaling activity. Genetically downstream or in parallel of glp-1 are the paralogous fbf-1 and fbf-2 genes, collectively termed fbf, which encode RNA binding proteins homologous to Drosophila Pumilio (Zhang et al. 1997). FBF directly represses gld-1 (Crittenden et al. 2002), synaptonemal complex genes (Merritt and Seydoux 2010), and likely some additional meiotic genes (Kershner and Kimble 2010), thereby promoting germline stem cell fate and/or inhibiting meiotic entry. FBF represses its direct messenger RNA (mRNA) targets by binding specific sequence sites within their 3′ UTR, resulting in translational inhibition and/or mRNA destabilization (Crittenden et al. 2002; Suh et al. 2009; Voronina et al. 2012). Additional genes downstream of glp-1 must also contribute to promoting germline stem cell fate, since fbf null mutants display a less-severe germline proliferation defect phenotype compared to glp-1 null mutants (Crittenden et al. 2002), and fbf mutants can maintain a stem cell pool when grown at higher temperatures (Merritt and Seydoux 2010; Hansen and Schedl 2013). Furthermore, how glp-1 signaling might promote fbf gene activity is unclear, although fbf-2 is proposed to be a direct glp-1 signaling target (Lamont et al. 2004). Recently, lst-1 and sygl-1 were identified as two direct transcriptional targets of glp-1 signaling that are redundantly required for germline stem cell proliferation and maintenance (Kershner et al. 2014), but how they act to promote the stem cell fate is not well understood.
GLD-1 protein expression levels are regulated during germline stem cell differentiation (Francis et al. 1995b; Jones et al. 1996). GLD-1 levels are low in germline stem cells and high in meiotic germ cells where GLD-1 is required for meiotic prophase progression/maintenance (Francis et al. 1995a; Ciosk et al. 2006). High GLD-1 is sufficient to drive premature meiotic entry of all germline stem cells (Crittenden et al. 2002; Hansen et al. 2004b), highlighting that repression of GLD-1 protein levels is one mechanism whereby glp-1 promotes germline stem cell fate and/or inhibits meiotic entry. GLD-1 accumulation is controlled primarily through post-transcriptional gene regulation, with much of its control mediated through the gld-1 3′ UTR (Jones et al. 1996; Merritt et al. 2008). FBF represses GLD-1 accumulation in stem cells (Crittenden et al. 2002; Suh et al. 2009; Voronina et al. 2012) but other unidentified repressors likely exist (Hansen et al. 2004b). gld-2 and gld-3 promote GLD-1 accumulation in addition to their other meiotic entry gene targets (Hansen et al. 2004b; Suh et al. 2006; Schmid et al. 2009; Nousch et al. 2014). Therefore, GLD-1 accumulation is likely tightly coupled to progression of germline stem cell differentiation and detailed understanding of its accumulation may provide new insight into glp-1-mediated control of the polarized pattern of germline stem cell differentiation.
Here we establish a sensitive assay to quantify GLD-1 levels across the field of developing cells of the proliferative zone in wild-type adult hermaphrodites (Figure 1) and compare levels to those in worms with known or postulated GLD-1 regulators. We find that glp-1 transcriptional targets lst-1 and sygl-1 are individually required for GLD-1 repression in stem cells. We also find that regulation of GLD-1 levels and pattern appears regional in the proliferative zone, which we model in relation to the current view of glp-1-mediated control of germline stem cell differentiation in the adult C. elegans hermaphrodite.
Materials and Methods
Nematode strain maintenance and genetics
All strains were maintained under standard growth conditions (Wood 1988) at 20°, unless noted otherwise, and null alleles were used in most cases, unless noted otherwise. All strain information is listed in Supporting Information, Table S1. Deletion mutants ok91 and ok224 are fbf-1 null alleles and behave similarly (Crittenden et al. 2002).
GLD-1 Quantification
Dissection and staining:
The following protocol was modified from other sources (Jones et al. 1996; Hansen et al. 2004a,b). A general outline is presented in Figure S1. An equal number of 24-hr past mid-L4 stage adult spo-11(ok79) (to be differentiated from test gonads, see Figure S1 and below) and the genotype to be quantified were transferred to the same 1.5-ml microcentrifuge tube of PBS for a total of ∼100–150 worms. Worms were then dissected and stained following the batch method (Francis et al. 1995a), where all dissected tissue was incubated within small glass tubes rather than on a slide. We believe this allows for more equivalent distribution of solutions across all tissue present in the sample and less variation within an individual experiment. Dissected gonads were fixed in 3% paraformaldehyde solution for 10 min at room temperature (21–23°) and then postfixed in cold (−20°) 100% methanol and stored at −20° for 30 min. Following multiple washes in PBS + 0.1% Tween-20, gonads were incubated overnight in affinity-purified rabbit-anti-GLD-1 antibodies (Jones et al. 1996) at 1:100 in 30% goat serum at room temperature. The same high concentration fractions were used for all experiments. Primary antibodies were removed, and gonads were washed with PBS + 0.1% Tween-20 three times for ∼10 min each wash. Gonads were then incubated in antirabbit Alexa Fluor 488 (Invitrogen) at 1:400 in 30% goat serum for ∼4 hr at room temperature. Gonads were washed three times in PBS + 0.1% Tween-20, with the final wash containing 100 ng/ml 4′,6-diamidino-2-phenylindole (DAPI) before mounted in a solution of ∼50% glycerol plus 1,4-diazabicyclo[2.2.2]octane (DABCO) onto a slide containing a freshly made 2.5% agarose pad.
Image acquisition:
Images were collected the same day the slide was prepared using a ×63 objective lens on a Zeiss Axioskop microscope equipped with a Hamamatsu camera using Axiovision software (Zeiss). Gonads were imaged individually and genotype was determined retrospectively as outlined in Figure S1B. Only unobstructed, nondamaged gonads were imaged. The distal end of 15 gonads per genotype on the slide were imaged for most experiments. Exposure time for image acquisition of GLD-1 stained gonads was constant within each experiment but varied between experiments (most were 30 ms, range 25–80 ms) and was determined using autoexposure in Axiovision software as a means of collecting pixel intensity levels within a linear range and to avoid pixel saturation. For each gonad, an image was collected for DAPI staining and for anti-GLD-1.
Image analysis:
DAPI and anti-GLD-1 images of the same gonad were stacked in ImageJ (National Institutes of Health), which allowed for a segmented line to be generated in the DAPI image and quantified in the GLD-1 image (Figure S1C). A line 35-cell diameter in length was manually drawn on each gonad that bisected the gonad using DAPI-stained nuclei to count each cell diameter. A line, 25-cell diameter in length, was used for genotypes where the start of meiotic entry (sME) was closer to the distal end, and a line, 45-cell diameter in length, was used for genotypes where the sME was further from the distal end (Table 1). The “line width” was set to 30 pixels in ImageJ (Figure S1). Plot profile was used to generate pixel intensity data across the length of the line. A custom Perl script (available upon request) was used to perform the following: Each x-coordinate from the plot profile line was divided by the length of the line, then multiplied by 35 to convert the x-coordinate to a value corresponding to cell diameter position. All pixel intensity data were pooled by integer and the average of all pixel values for a given cell diameter was used as the representative value for an individual gonad. All cell diameter values within an experiment were normalized by setting the cell diameter with the highest average value for spo-11(ok79) gonads to 100, conforming to the “relative GLD-1 levels” scale.
Table 1. Summary of proliferative zone size and cell diameter distance to meiotic entry region.
| Genotype | Proliferative zone GCsa mean ± SD (range) | sMEb mean ± SD (range) | eMEc mean ± SD (range) | n |
|---|---|---|---|---|
| Wild type | 246 ± 27 (201–314) | 19 ± 2 (16–23) | 26 ± 2 (22–31) | 30 |
| gld-1 repression | ||||
| fbf-1(ok91) | 226 ± 20 (194–266) | 15 ± 2 (12–17)* | 22 ± 2 (19–24)* | 15 |
| glp-1(bn18) | 144 ± 23 (117–191)* | 10 ± 2 (7–13)* | 15 ± 3 (12–20)* | 15 |
| fbf-1(ok224); glp-1(bn18) | 134 ± 15 (116–159)* | 9 ± 2 (6–12)* | 15 ± 3 (9–20)* | 15 |
| fbf-2(q738) | 296 ± 21 (238–324)* | 23 ± 3 (19–28)* | 31 ± 3 (26–37)* | 15 |
| gld-1 activation | ||||
| gld-2(q497) | 339 ± 37 (265–393)* | 21 ± 2 (17–26)* | 34 ± 4 (25–42)* | 15 |
| gld-3(q730) | 283 ± 26 (243–320)* | 22 ± 3 (15–29)* | 28 ± 4 (24–40)* | 15 |
| gld-3(q741) | 333 ± 38 (275–430)* | 20 ± 3 (15–23)* | 28 ± 3 (20–35)* | 15 |
| nos-3(oz231) | 176 ± 18 (143–203)* | 15 ± 2 (11–20)* | 23 ± 3 (19–28)* | 15 |
| Physiological effect | ||||
| eat-2(ad465) | 110 ± 15 (85–139)* | 12 ± 3 (7–17)* | 16 ± 2 (12–23)* | 15 |
| eat-5(ad464) | 160 ± 35 (106–211)* | 16 ± 2 (11–21)* | 22 ± 3 (17–28)* | 15 |
| daf-1(m40)d | 154 ± 20 (120–185)* | 16 ± 2 (12–20)* | 22 ± 2 (19–25)* | 15 |
| Putative regulators | ||||
| lst-1(ok814) | 183 ± 19 (140–205)* | 17 ± 2 (13–21)* | 22 ± 2 (16–25)* | 15 |
| sygl-1(tm5040) | 137 ± 15 (107–168)* | 12 ± 2 (9–16)* | 17 ± 2 (14–21)* | 15 |
| mir-35 fam(0)e | 186 ± 35 (130–252)* | 17 ± 3 (12–20)* | 23 ± 3 (18–26)* | 15 |
SD, standard deviation.
Significantly different than wild type, P-value ≤0.01, Fisher’s uncorrected LSD.
GC, germ cells, proliferative zone cell is considered REC-8 positive, HIM-3 negative nucleus.
sME, start of meiotic entry region, number of germ cell diameters from distal end to the first HIM-3 positive germ cell.
eME, end of meiotic entry region, number of germ cell diameters from distal end to the last REC-8 positive germ cell.
Strain maintained at 15°, shifted to 20° at mid-L4 stage.
Null(0) for mir-35 family; actual genotype mir-35 thru 41(nDf50) mir-42-45(nDf49).
We note that the GLD-1 raw pixel intensity value accumulation curves can differ substantially between gonads of the same genotype within an experimental replicate, and we attribute much of the variation to “biological” noise. Thus, to allow conclusions to be drawn from the data, we employed large sample sizes, a minimum of three replicates, and a stringent statistical threshold (P = 0.01) for comparison of means as follows: All statistical analysis was performed with data converted to the relative GLD-1 levels scale. A minimum of three replicate experiments was pooled for a total of ∼45 or more data points per cell diameter per genotype. The pooled data for a given genotype at base, sME, and peak were used in a one-way ANOVA followed by a multiple comparison test for differences from wild-type levels using Fisher’s least significant difference (LSD) test, with a P-value of 0.01 or less considered significantly different from wild type. The statistical analysis was performed using Prism 6 (GraphPad Software).
GLD-1 levels scale:
For simplicity, relative GLD-1 level values were converted to GLD-1 levels by subtracting all individual values of each genotype by the mean relative GLD-1 level values obtained for gld-1(q485), a gld-1 protein null (Jones et al. 1996), followed by transforming all data so the mean wild-type peak value equaled 100. The GLD-1 level scale then corresponds to percentage of wild-type peak GLD-1 level.
Representative images:
Images of GLD-1 staining were selected by identifying individual gonads with GLD-1 level values close to the mean levels identified for a given genotype. The image was processed in ImageJ and Photoshop (Adobe) and individual gonads were cut from their original source and placed on a black background to fit within a smaller field of view.
GLD-1::GFP quantification
Dissection and fixation were performed as described for GLD-1 quantification, except that incubation time in 3% paraformaldehyde solution was for 5 min and no postfix with methanol was used. After fixation, dissected worms/gonads were washed three times in PBS + 0.1% Tween-20 then incubated in PBS + 0.1% Tween-20 and DAPI (100 ng/µl final concentration) for >5 min prior to mounting onto slides with DABCO solution. Slides were imaged immediately after preparation.
Image acquisition
All GFP images were taken at 350-ms exposure time. For an experimental replicate, 15 ozIs5[gld-1::gfp] (Table S1; Schumacher et al. 2005) distal gonads were imaged along with 15 spo-11(ok79) gonads, which served as a fluorescence background control. All image processing was performed as described for quantification of GLD-1 levels by antibody staining. Three biological replicates were performed.
Corrected GFP intensity
The mean intensity value by germ cell diameter calculated for spo-11(ok79) from the pooled data was used as a baseline value and subtracted from all individual values collected for GLD-1::GFP. Each individual data point was then transformed so the largest mean value obtained in GLD-1::GFP equaled 100.
gld-1 3′ UTR GFP reporter quantification
Dissection, fixation, and staining of axIs1723 (Table S1; Merritt et al. 2008) were performed as described for GLD-1, except affinity purified rabbit anti-GFP antibodies (kind gift from S. Arur, University of Texas MD Anderson Cancer Center, Houston) were used at 1:100 dilution. Secondary antibodies used were antirabbit IgG-conjugated Alexa Fluor 594 (Invitrogen) at 1:400 dilution. Image acquisition was performed as described for GLD-1 immunofluorescence with 100-ms exposure time in two replicate experiments. GFP quantification was performed as described above for GLD-1::GFP quantification.
Modeling GLD-1 accumulation pattern as a sigmoid curve
The relative GLD-1 intensity values were background corrected similar to the GLD-1 levels scale. These background-corrected values were then modeled by nonlinear regression analysis with the following settings: 4PL-dose response curve, data weighted (1/Y2) in Prism 6 (GraphPad Software). Data were weighted to account for the difference in standard deviations at base vs. sME and peak. The background-corrected values for each model were transformed so the top value calculated for wild type equaled 100. Steepness values from each genotype were compared pairwise with wild type using goodness-of-fit analysis and considered different based on a threshold P-value of 0.01. Corrected GFP intensity values were used to model data for GLD-1::GFP and gld-1 3′ UTR reporter GFP.
Proliferative zone counts and position of start and end of meiotic entry
Immunofluorescence of REC-8 and HIM-3:
Immunofluorescence of REC-8 and HIM-3 were performed essentially as described elsewhere (Hansen et al. 2004a; Fox et al. 2011). Dissected gonads were fixed in 3% paraformaldehyde solution for 10 min at room temperature and then postfixed with 100% methanol for 1 hr (REC-8/HIM-3) at −20°. Incubations were performed as for GLD-1 except primary antibodies were: rat-anti-REC-8 (Pasierbek et al. 2001) (1:100), and mouse-anti-HIM-3 (Zetka et al. 1999) (1:100). Secondary antibodies were: antirabbit IgG-conjugated Alexa Fluor 488 and antirat IgG-conjugated Alexa Fluor 594 (Invitrogen). Slides were prepared as for GLD-1 staining.
Image acquisition for REC-8/HIM-3 staining:
Images were collected with a ×63 objective lens on a PerkinElmer spinning disc confocal microscope using Volocity software. Approximately 20, 1-µm separated z-plane images were acquired for each gonad. The number of gonads imaged per genotype is listed in Table 1.
Proliferative zone counts:
Counts were performed manually in ImageJ using the Cell Counter plug-in. A REC-8 positive, HIM-3 negative nucleus was considered a proliferative zone cell (Table 1). Representative images of REC-8/HIM-3 staining were processed in ImageJ and Photoshop. Gonad images were cut from their original source and placed on a black background to fit within a smaller field of view.
Results
Comparative quantification of GLD-1 levels
GLD-1 is a cytoplasmic protein whose levels are low in germline stem cells in the distal-most region of the C. elegans adult hermaphrodite gonad and high in meiotic germ cells (Jones et al. 1996). However, quantitative data are necessary to determine the contributions of individual regulators and to assess the contribution of potential regulators. The gonad is readily isolated from the rest of the worm via dissection, allowing GLD-1 levels to be quantified by immunofluorescence, following anti-GLD-1 antibody staining.
A number of considerations were taken to quantify GLD-1 by immunofluorescence (Waters 2009; Wolf et al. 2013). We reasoned that a large source of fluorescence intensity variation would be from comparing GLD-1 levels between genotypes, since each genotype is processed in separate immunofluorescence experiments owing to similar gonad morphology after dissection. To overcome this variation, we first used gonads from spo-11(ok79) worms as an internal normalization control for fluorescent intensity (Figure S1). spo-11(ok79) gonads had normal GLD-1 levels compared to wild type (Figure S2) and a readily observable diakinesis morphology phenotype (Dernburg et al. 1998) to distinguish spo-11 mutant gonads from experimental gonads after dissection and staining (Figure S1B). The use of spo-11(ok79) as an internal control reduced variation over other means of normalization (see Figure S2). Second, we pooled a minimum of three biological replicates, where ∼15 gonads per genotype were quantified per replicate and utilized a high statistical threshold (P = 0.01) to call relevant differences.
The quantified mean distribution of GLD-1 for wild-type adult hermaphrodites across the distal 35 germ cell diameters (gcd) is in Figure 1D. GLD-1 levels were lowest in the first gcd and then elevated to a peak level ∼23–28 gcd from the distal end. The peak level of GLD-1 corresponded to the position where essentially all germ cells had entered meiosis, as determined by REC-8/HIM-3 staining, which are established markers of the proliferative zone (Hansen et al. 2004a; Fox et al. 2011) (see Figure 1). In wild type, the average sME—position in gcd from the distal end where germ cells first stain positive for HIM-3—occurred at ∼19 gcd and end of the meiotic entry region (eME)—position in gcd, where the most proximal germ cells stain positive for REC-8—occurred at ∼26 gcd from the distal end (Table 1). GLD-1 levels were below peak at sME (Figure 1D), indicating GLD-1 peaks when most germ cells have entered meiotic prophase.
We compared GLD-1 levels at three biologically relevant positions defined as base, sME, and peak (Figure 1D). Base is the GLD-1 level at the distal-most gcd, sME is the GLD-1 level at the average sME for a given genotype, and peak is the GLD-1 level at the average eME for a given genotype (see Table 1).
A large source of background fluorescence signal is from the tissue itself. Therefore, we compared fluorescence intensity of wild type to gld-1(q485) (Figure S3, B, D, and F–H), an RNA null allele of gld-1 (Francis et al. 1995a; Jones et al. 1996). Base was higher in wild type compared to gld-1(q485) (Figure S3F), indicating that GLD-1 is present and detectable on average in the distal-most gcd. Once rescaled to control for background signal intensity and transformed so that mean wild-type peak levels equaled 100 (see Materials and Methods), base GLD-1 levels in wild type were ∼5% of peak (Table S2) and GLD-1 levels change ∼20-fold in the transition from stem cell to leptotene/zygotene (Table S3).
We also quantified GFP levels from a strain of worms expressing a genomic GLD-1 transgene conjugated to GFP (gld-1::gfp) (ozIs5; Schumacher et al. 2005) as an additional means of quantifying GLD-1 levels (Figure S4A). GFP signal gives a qualitatively similar accumulation curve over the distal-most 30 gcd (compare Figure 1D to Figure S4A). Quantification of GFP expression was comparable to our results with GLD-1 immunofluorescence; GLD-1::GFP appeared detectable at base and GLD-1::GFP elevated ∼30-fold from base to peak levels (Figure S4, A and,C). However, we found that background signal was elevated when quantifying GLD-1::GFP, thereby adversely reducing signal to noise compared to GLD-1 immunofluorescence. Therefore, we concluded that quantifying GLD-1 by immunofluorescence would produce a more sensitive assay to compare GLD-1 levels between genotypes.
Many germline-expressed genes are regulated at a post-transcriptional level through their 3′ UTRs (Merritt et al. 2008), including gld-1. To determine the quantitative contribution of 3′ UTR regulation to posttranscriptional regulation of GLD-1 accumulation, we quantified GFP immunofluorescence in gonads from worms with a gld-1 3′ UTR reporter GFP (Figure S4B). Similar accumulation curves were observed in the distal-most 20 gcd compared to GLD-1 immunofluorescence (compare Figure 1D to Figure S4B). We estimated that GFP levels elevated ∼30-fold in gonads from worms with the gld-1 3′ UTR reporter GFP transgene axIs1723 (Table S1; Figure S4B; Merritt et al. 2008), suggesting that most of the distal GLD-1 accumulation pattern is quantitatively controlled by its 3′ UTR. However, we note a very high level of variation of GFP accumulation between worms with the gld-1 3′ UTR reporter, which may be a property of the transgene and/or integration site. Therefore, we concluded that quantifying GLD-1 by immunofluorescence would produce a more sensitive assay to compare GLD-1 levels between genotypes.
Low GLD-1 base levels require genes involved in gld-1 repression
Genes known to be involved in repression of GLD-1 accumulation include fbf-1, fbf-2, and glp-1 (Crittenden et al. 2002; Hansen et al. 2004b) and we examined worms lacking or mutant in these genes for changes in GLD-1 levels (Figure 2). Since FBF-1 and FBF-2 are present in germ cells from the distal-most germ cell through sME/eME (Lamont et al. 2004; Voronina et al. 2012), we expected their loss would result in elevation of GLD-1 throughout the distal germline. Instead, fbf-1(ok91) mutants had elevated base GLD-1 levels relative to wild type (Figure 2, B, F, and J), but sME and peak GLD-1 levels were no different than wild type (Figure 2, B, F, K, and L). Therefore, fbf-1 represses GLD-1 levels predominately in distal germ cells, but not in meiotic germ cells.
Figure 2.
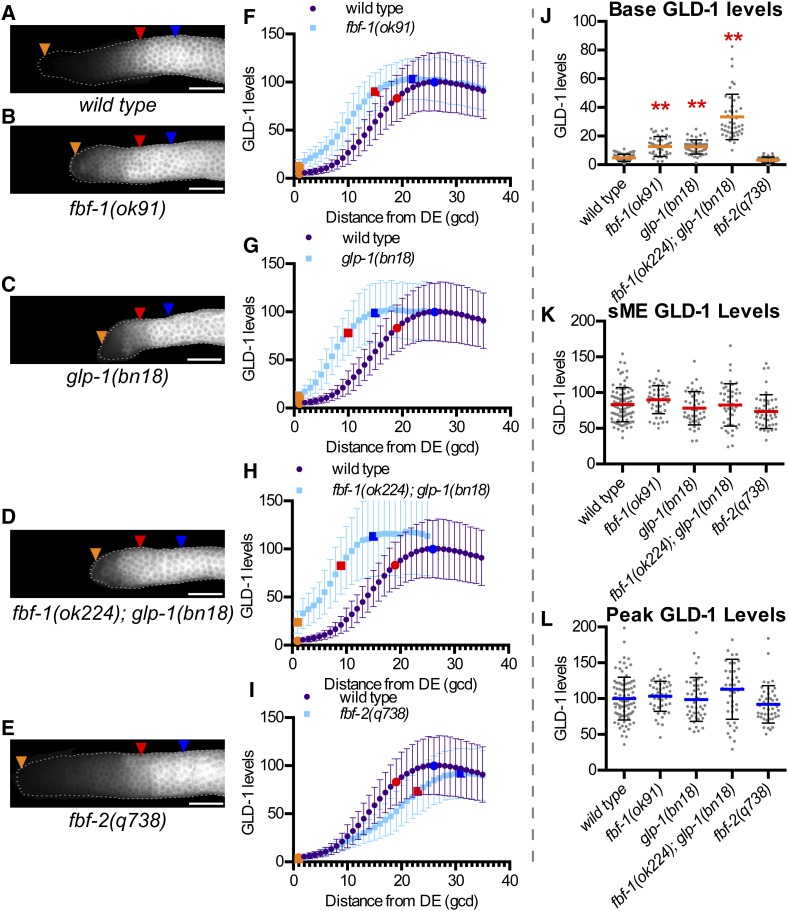
Genes involved in repressing GLD-1 accumulation are required for low GLD-1 base. (A–E) Representative GLD-1 immunofluorescence images for the indicated genotypes. Orange triangles, 1st gcd; red triangles, sME; blue triangles, eME; and white bars are 25 µm. (F–I) Plots of GLD-1 levels. DE, distal end. Error bars ± SD. Orange dot, base; red dot, sME; blue dot, peak. Scatterplots are of (J) base, (K) sME, (L) peak. **P ≤ 0.01, Fisher’s LSD, in red, mean is higher than wild type.
To evaluate glp-1 signaling in controlling GLD-1 levels, we used the bn18 temperature-sensitive allele that results in all proliferative zone cells entering meiosis at restrictive temperature (25°) (Kodoyianni et al. 1992); at permissive temperatures (15 or 20°), glp-1 activity is apparently reduced (Fox and Schedl 2015). Consistent with reduced glp-1 activity, base GLD-1 level was elevated in glp-1(bn18) at 20° compared to wild type (Figure 2, C, G, and J). sME and peak GLD-1 level were not different (Figure 2, C, G, K, and L). Thus, glp-1 controls GLD-1 levels in the distal-most germ cells but not in germ cells entering meiosis, consistent with our expectation of localized glp-1 signaling activity.
Loss of fbf-1 and reduced glp-1 activity each resulted in elevation of base GLD-1 level to ∼12–13% of peak, compared to ∼5% in wild type (Table S2). Since a proliferative pool is maintained in both genotypes, 13% of peak is not sufficient to commit all proliferative fate germ cells to meiosis. We tested if further elevated GLD-1 in fbf-1(ok224); glp-1(bn18) mutants was sufficient to cause all distal germ cells to enter meiosis, but essentially all fbf-1(ok224); glp-1(bn18) mutants maintained a proliferative pool at 20° (Table 1, Figure S5). Base GLD-1 level in fbf-1(ok224); glp-1(bn18) was increased relative to the single mutants (Figure 2, D, H, and J), reaching ∼33% of peak (Table S2); however, this level is not sufficient to prematurely drive all germ cells into meiosis. sME and peak GLD-1 levels in fbf-1(ok224); glp-1(bn18) were no different than wild type (Figure 2, D, H, K, and L), reinforcing that repression of GLD-1 accumulation by fbf-1 and glp-1 is limited to distal-most germ cells.
GLD-1 levels in fbf-2(q738) at base, sME, and peak were no different than wild type (Figure 2, E, I, and J–L), indicating that fbf-2 is individually dispensable for normal GLD-1 levels. fbf-2(q738) mutants maintain a larger proliferative zone than wild type (Table 1), due in part to modestly elevated and ectopic FBF-1 (Lamont et al. 2004; Voronina et al. 2012), potentially masking an individual requirement for FBF-2 in repression of GLD-1 levels.
High GLD-1 peak levels require genes involved in gld-1 activation
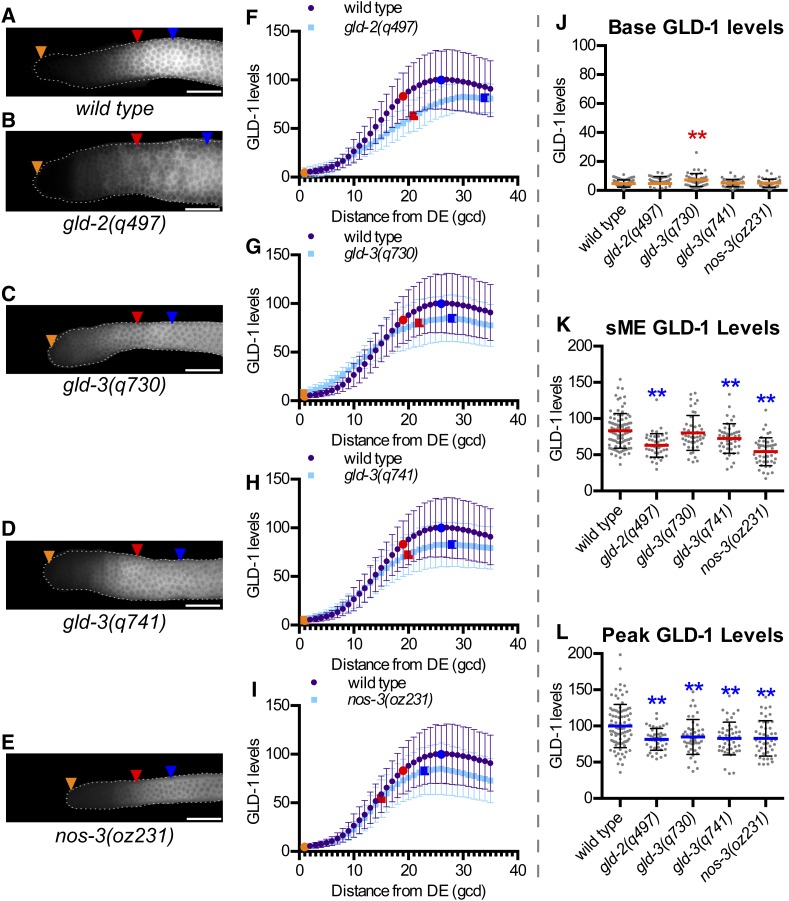
Next we quantified GLD-1 levels in mutants of genes known to promote activation of GLD-1 accumulation: gld-2, gld-3, and nos-3 (Hansen et al. 2004b; Suh et al. 2006) (Figure 3), which were expected to have reduced GLD-1 levels. sME and peak GLD-1 levels were lower in gld-2(q497) compared to wild type (Figure 3, B, F, K, and L), consistent with gld-2 promoting high GLD-1 levels as germ cells enter meiosis. In contrast, base in gld-2(q497) was similar to wild type (Figure 3, B, F, and J), suggesting that gld-2 may not promote GLD-1 levels in the distal-most germ cells.
Figure 3.
Genes involved in activating GLD-1 accumulation are required for high GLD-1 peak. (A–E) Representative GLD-1 immunofluorescence images for the indicated genotypes. Orange triangles, 1st gcd; red triangles, sME; blue triangles, eME; and white bars are 25 µm. (F–I) Plots of GLD-1 levels. DE, distal end. Error bars ± SD. Orange dot, base; red dot, sME; blue dot, peak. Scatterplots are of (J) base, (K) sME, (L) peak. **P ≤ 0.01, Fisher’s LSD, in red, mean is higher than wild type; in blue, mean is lower than wild type.
gld-3(q730) mutants had lower peak compared to wild type (Figure 3, C, G, and L), consistent with gld-3 acting to promote high GLD-1 levels as germ cells enter meiosis. Surprisingly, gld-3(q730) mutants had a higher base GLD-1 level than wild type (Figure 3, C, G, and J), implicating gld-3 in repression of GLD-1 levels in the distal-most germ cells. We further explored the potential repressive and activating functions of gld-3 using the gld-3(q741) allele, which produces GLD-3S and a truncated version of GLD-3L (Eckmann et al. 2004). gld-3(q741) mutant gonads had similar base GLD-1 levels as wild type (Figure 3, D, H, and J), while peak in gld-3(q741) was reduced relative to wild type (Figure 3, D, H, and L). One interpretation of this result is that GLD-3S is sufficient for gld-3 repression of GLD-1 levels, while GLD-3L is required for gld-3 promotion of high GLD-1 peak levels. GLD-3S may also promote peak, but does not appear sufficient. Confounding interpretation of GLD-1 levels in q741 is that a truncated form of GLD-3L is also formed in these mutant animals, which may display gain-of-function properties instead of a true GLD-3L null. Further complicating GLD-3S as a repressor of GLD-1 base levels is that the gld-3S mRNA is proposed to be regulated by FBF but not the gld-3L mRNA (Eckmann et al. 2004), which is counterintuitive for a gene that acts to repress GLD-1.
Lastly, we quantified GLD-1 in nos-3(oz231) and found that both sME and peak were reduced in nos-3(oz231) compared to wild type (Figure 3, E, I, K, and L), consistent with nos-3 acting to promote high GLD-1 levels in germ cells entering meiosis. Base GLD-1 level in nos-3(oz231) was similar to wild type (Figure 3, E, I, and J), indicating that nos-3 is not required for basal expression in the distal-most germ cells.
GLD-1 levels are insensitive to gld-1 gene dose
A common approach to reduce gene activity is to examine animals heterozygous for a null allele, thereby reducing gene dose by half. Worms heterozygous for a null allele of gld-1 [gld-1(q485)/+] display phenotypes consistent with reduced gld-1-gene dose, including some animals displaying a germline feminization phenotype (Francis et al. 1995b) and gld-1(q485)/+; fbf-1fbf-2 mutants do not display premature meiotic entry defects like those of fbf-1fbf-2 mutants (Crittenden et al. 2002). Surprisingly, we observed that gld-1(q485)/+ adults had wild-type GLD-1 levels (Figure S3, C and E–H). The overall pattern of GLD-1 and size of the proliferative zone in gld-1(q485)/+ were essentially identical to wild type (Figure S3). Both the germline feminization phenotype and the premature meiotic entry defects of fbf-1fbf-2 double mutants occur during larval development and are sensitive to gld-1 gene dose. The normal GLD-1 levels in gld-1(q485)/+ adults could suggest a feedback mechanism in the adult germline that compensates for the reduced gene dose.
GLD-1 base and peak are unchanged by loss of genes involved in physiological regulation of germline size
Nonoptimal physiological conditions can result in reduced adult proliferative zone sizes (Hubbard et al. 2013). We wondered if this might be mediated through elevation of GLD-1 levels, analogous to glp-1(bn18). To explore this idea, we examined worms lacking genes that influence physiological regulation of proliferative zone size (eat-2, eat-5, daf-1, and daf-2) (Figure 4) where their roles in physiology and reduction of proliferative zone sizes are largely independent of the glp-1 pathway (Michaelson et al. 2010; Korta et al. 2012; Dalfó et al. 2012) (Table 1).
Figure 4.
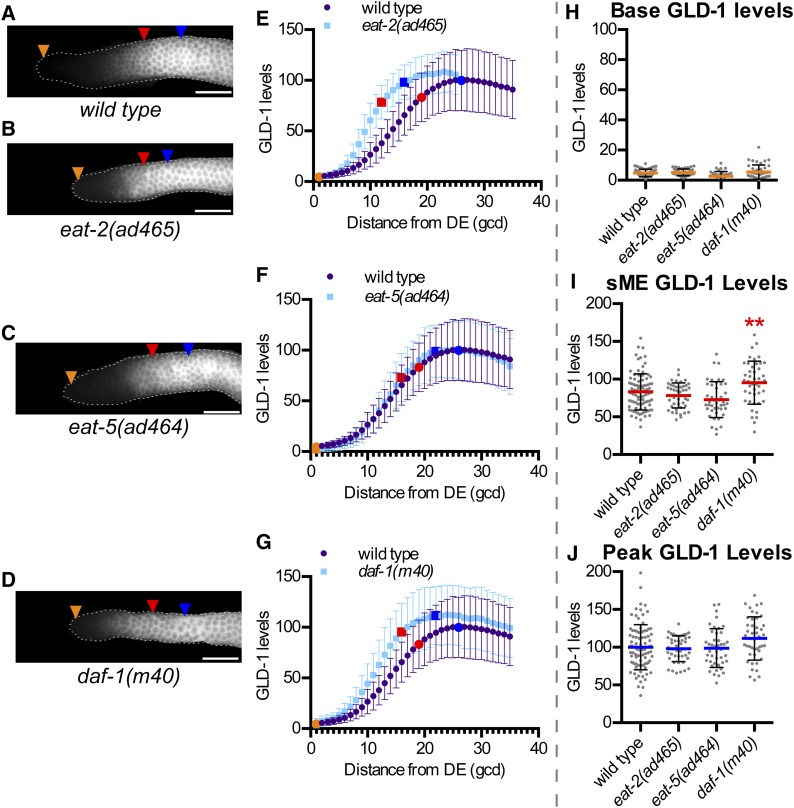
Genes involved in physiological regulation of germline size do not affect GLD-1 levels. (A–D) Representative GLD-1 immunofluorescence images for the indicated genotypes. Orange triangles, 1st gcd; red triangles, sME; blue triangles, eME; and white bars are 25 µm. (E–G) Plots of GLD-1 levels. DE, distal end. Error bars ± SD. Orange dot, base; red dot, sME; blue dot, peak. Scatterplots are of (H) base, (I) sME, (J) peak. **P ≤ 0.01, Fisher’s LSD, in red, mean is higher compared to wild type. Elevated sME in daf-1(m40) may reflect a subtle delay between elevation of GLD-1 and execution of the start of meiotic prophase.
eat-2(ad465) mutants, which are less efficient at eating food due to impaired pharyngeal pumping (Avery 1993) and consequently have smaller proliferative zones than wild type (Korta et al. 2012) (Table 1), had GLD-1 levels similar to wild type (Figure 4, B, E, H–J). eat-5(ad464), which has less severe pleiotropic phenotypes compared to eat-2 but still smaller than wild-type proliferative zones (Table 1), had GLD-1 levels similar to wild type (Figure 4, C, F, and H–J). Therefore, reduced food intake, while initiating a physiological response that reduces proliferative zone size, does not influence GLD-1 levels at base, sME, or peak.
Physiological changes associated with environmental stimuli are transmitted to the germline through multiple signaling pathways (Hubbard et al. 2013). The TGF-β signaling pathway functions germ-cell nonautonomously to influence proliferative zone size (Dalfó et al. 2012). Mutants in daf-1, a TGF-β signaling receptor (Georgi et al. 1990), had base and peak GLD-1 levels that were no different than wild type (Figure 4, D, G, H, and J). Insulin-like signaling also influences proliferative zone size (Michaelson et al. 2010), but our quantification of GLD-1 in daf-2 mutants (Figure S6) did not support a role for this pathway in controlling GLD-1 levels. Therefore, impaired physiological signaling induces a response to reduce proliferative zone size, but does not alter GLD-1 levels at base or peak.
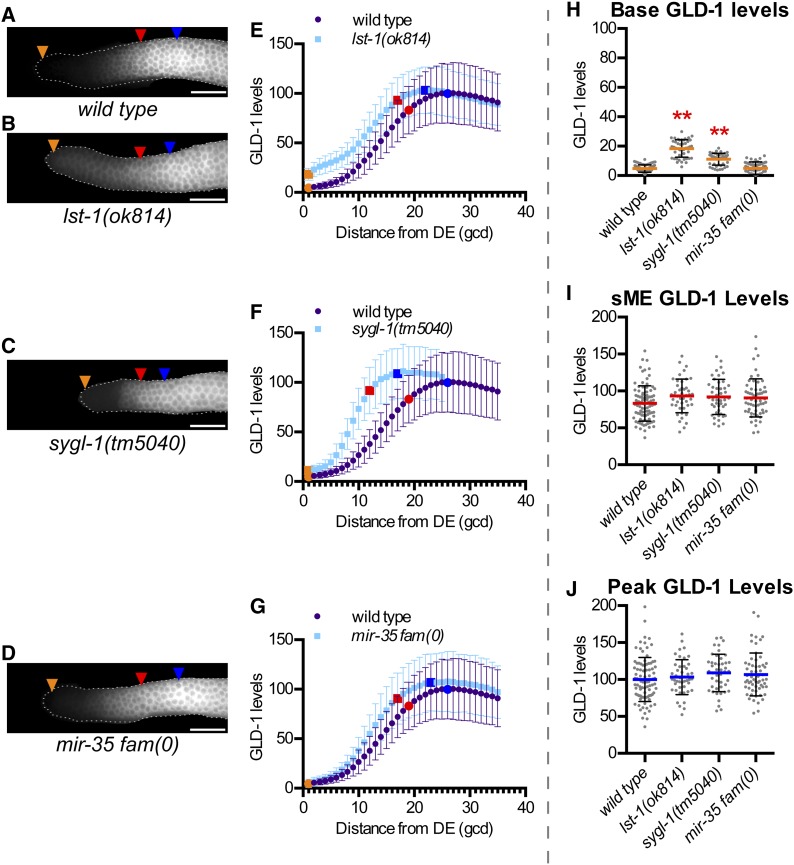
Low GLD-1 base levels require lst-1 and sygl-1, but not the mir-35 family
We next examined two genes and a gene family for possible roles in regulation of GLD-1 accumulation: lst-1, sygl-1, and the mir-35 family of microRNAs (Figure 5). glp-1 signaling represses GLD-1 accumulation (Hansen et al. 2004b), but the identity of direct transcriptional targets of glp-1 that mediate repression of GLD-1 are unknown. lst-1 and sygl-1 are direct transcriptional targets of the glp-1 signaling pathway that are redundantly required for germline stem cell fate and act upstream of the gld-1 and gld-2 pathways (Kershner et al. 2014). lst-1 and sygl-1 might act to repress GLD-1 levels. Both lst-1(ok814) and sygl-1(tm5040) mutants had elevated base GLD-1 levels compared to wild type (Figure 5, B, C, E, F, and H). sME and peak GLD-1 levels in both lst-1(ok814) and sygl-1(tm5040) mutants were no different than wild type (Figure 5, B, C, E, F, I, and J), indicating lst-1 and sygl-1 influence GLD-1 levels specifically in distal-most germ cells, consistent with their regulation by glp-1 signaling.
Figure 5.
lst-1 and sygl-1, but not mir-35 family, are required for low GLD-1 base levels. (A–D) Representative GLD-1 immunofluorescence images for the indicated genotypes. mir-35 fam(0) corresponds to mir-35 through 41(nDf50) mir-42-45(nDf49). Orange triangles, 1st gcd; red triangles, sME; blue triangles, eME; and white bars are 25 µm. (E–G) Plots of GLD-1 levels. DE, distal end. Error bars ± SD. Orange dot, base; red dot, sME; blue dot, peak. Scatterplots are of (H) base, (I) sME, and (J) peak. **P ≤ 0.01, Fisher’s LSD, in red, mean is higher compared to wild type.
FBF-1/-2 post-transcriptionally repress GLD-1 levels to promote the germline stem cell fate (Crittenden et al. 2002). lst-1 and/or sygl-1 may act in the same pathway as fbf-1/-2 to promote the stem cell fate, or in parallel. lst-1(ok814); fbf-1(ok91) fbf-2(q704) and sygl-1(tm5040); fbf-1(ok91) fbf-2(q704) mutants produced fewer total and later stage meiotic germ cells than fbf-1(ok91) fbf-2(q704) mutants in late L4 (Table 2), consistent with enhanced premature meiotic defects. Therefore, lst-1 and sygl-1 act, at least partly, in parallel to fbf-1/2 to promote germline stem cell fate.
Table 2. lst-1 and sygl-1 enhance premature meiotic entry defect of fbf-1 fbf-2 mutants.
| Genotype | Proliferative zone GC no. (mean ± SD) | Pachytene or 1° spermatocyte (mean ± SD) | Spermatids (mean ± SD) | Total no. of GCs (mean ± SD)a | nb |
|---|---|---|---|---|---|
| Wild type | 180 ± 15 | 107 ± 34 | 0 | 286 ± 41 | 5 |
| fbf-1(ok91) fbf-2(q704) | 0 | 85 ± 16 | 2 ± 5 | 85 ± 16 | 10 |
| lst-1(ok814); fbf-1(ok91) fbf-2(q704) | 0 | 8 ± 25 | 73 ± 34 | 26** ± 19 | 10 |
| sygl-1(tm5040); fbf-1(ok91) fbf-2(q704) | 0 | 23 ± 24 | 63 ± 71 | 39** ± 10 | 10 |
The shift to more mature gametes in lst-1; fbf-1 fbf-2 and sygl-1; fbf-1 fbf-2 compared to fbf-1 fbf-2 is further indicative of earlier meiotic entry. SD, standard deviation.
P < 0.01 compared to fbf-1(ok91) fbf-2(q704), Sidak’s corrected multiple comparison test.
Sum of all germ cells counted. Each spermatid counted as 1/4 germ cell since 4 arise from a single 1° spermatocyte.
Number of individual gonads scored at late-L4 stage.
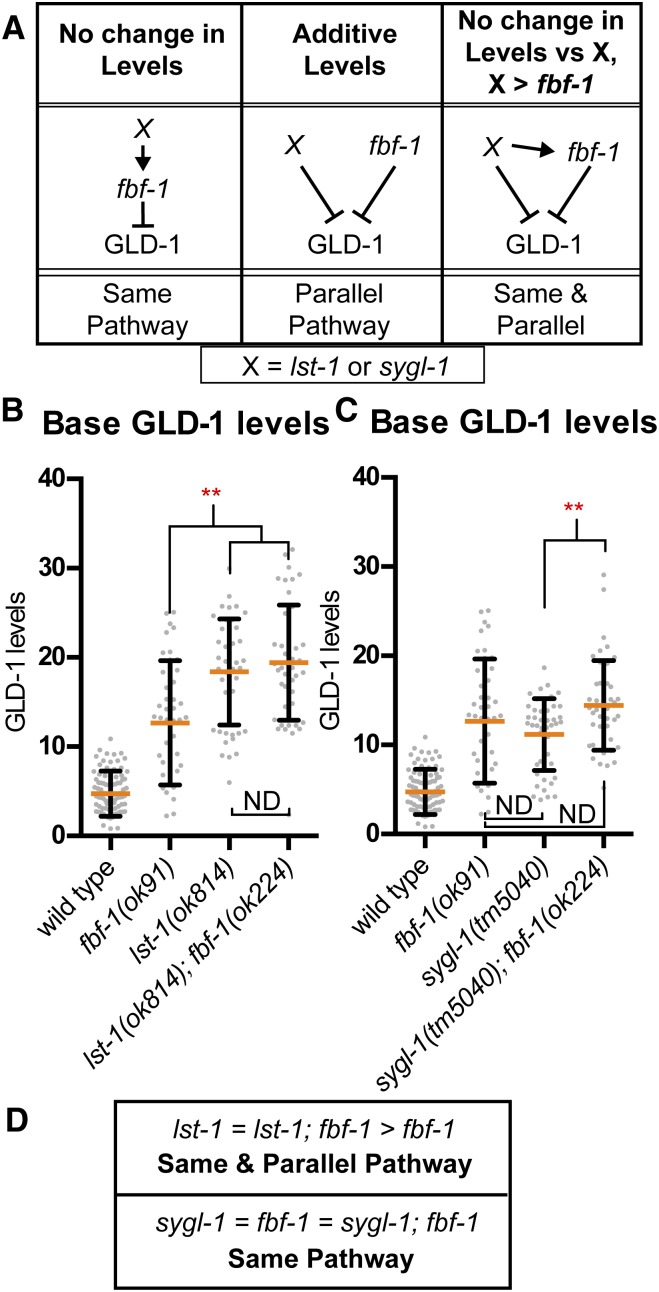
The severity of the meiotic entry phenotype in lst-1(ok814); fbf-1(ok91) fbf-2(q704) and sygl-1(tm5040); fbf-1(ok91) fbf-2(q704) mutants precluded our ability to measure GLD-1 levels in these strains. Instead, we quantified GLD-1 levels in lst-1(ok814); fbf-1(ok224) and sygl-1(tm5040); fbf-1(ok91) to determine if lst-1 or sygl-1 act in the same pathway or in parallel to fbf-1 to repress GLD-1 levels (Figure 6A). Base GLD-1 level in lst-1(ok814) was higher than in fbf-1(ok91) (Figure 6B), suggesting that lst-1 acts in parallel to fbf-1 to repress GLD-1 levels. In contrast, base GLD-1 level in lst-1(ok814); fbf-1(ok224) was no higher than in lst-1(ok814) (Figure 6B), indicating that lst-1 also acts in the same pathway as fbf-1 to repress base GLD-1 levels (Figure 6, A and D). We conclude that lst-1 acts in both the same pathway and in parallel to fbf-1 to repress GLD-1 levels. sygl-1(tm5040); fbf-1(ok224) mutants had base GLD-1 level that was no different from fbf-1(ok91), although it was higher than sygl-1(tm5040) (Figure 6C). However, base in sygl-1(tm5040) mutants were no different than in fbf-1(ok91) (Figure 6C). The simplest explanation of these data is that sygl-1 acts in the same pathway as fbf-1 to repress base GLD-1 levels (Figure 6D). Our observation that base GLD-1 levels in sygl-1(tm5040); fbf-1(ok224) were elevated relative to sygl-1(tm5040) may indicate additional complexity of GLD-1 repression by sygl-1. Nonstatistical comparison of the mean values shows a trend in GLD-1 base levels between the mutants as follows: sygl-1 mutants < fbf-1 mutants < sygl-1; fbf-1 double mutants, which might suggest small nonoverlapping activity between sygl-1 and fbf-1 in repressing GLD-1 base. Nonetheless, the small differences (in most comparisons not of statistical significance) in means between sygl-1 single mutants, fbf-1 single mutants, and sygl-1; fbf-1 double mutants suggests that any nonoverlapping activity is relatively minor, and instead sygl-1 likely predominately acts in the same pathway as fbf-1 to repress GLD-1 levels.
Figure 6.
lst-1 and sygl-1 act in fbf-1 pathway, and lst-1 also acts in parallel to repress GLD-1 base levels. (A) Predictions of double mutant phenotype for a gene acting in the same pathway as fbf-1 (left), parallel to fbf-1 (middle), or both (right). Scatterplots are of GLD-1 base for (B) lst-1 and (C) sygl-1 analysis. Red, **P ≤ 0.01, Fisher’s LSD, mean is higher compared to wild type. ND, not different. (D) Summary of observed outcomes for lst-1 and sygl-1 in relation to fbf-1 pathway in repressing GLD-1 levels.
Since our data indicate that lst-1 and sygl-1 act through fbf-1 to repress GLD-1 base levels, we further probed if fbf-1 singularly required lst-1 or sygl-1 for activity by generating mutants of lst-1 or sygl-1 with fbf-2. While fbf-1fbf-2 double mutants display premature meiotic entry defects, neither lst-1(ok814); fbf-2(q738) mutants or sygl-1(tm5040); fbf-2(q738) mutants displayed any severe premature meiotic entry defects (J. L. Brenner and T. Schedl, unpublished results). Since repression of GLD-1 levels is a major mechanism whereby fbf-1 represses meiotic entry, then lst-1 and sygl-1 likely redundantly promote fbf-1 activity to repress GLD-1.
The mir-35 family of microRNAs consists of eight microRNAs (mir-35 through mir-42) that were proposed to negatively regulate gld-1 to promote proliferative zone size (Liu et al. 2011), however GLD-1 levels were not examined. mir-35-41(nDf50) mir-42-44(nDf49) mutants [mir-35 fam(0)] had smaller proliferative zone sizes compared to wild type (Table 1), consistent with a potential role in repressing GLD-1 levels. However, GLD-1 levels in mir-35 fam(0) were no different than wild-type GLD-1 levels at base, sME, or peak (Figure 5, D, G, and H–J). Redundancy in repression of GLD-1 levels may mask an individual role for the mir-35 family. To explore this further, we examined worms mutant for the mir-35 family and genes known to repress GLD-1 (Figure S5 and J. L. Brenner and T. Schedl, unpublished results). fbf-1(ok224); mir-35 fam(0); glp-1(bn18) failed to show premature meiotic entry defects consistent with failure to repress GLD-1 (Figure S5B) and GLD-1 levels were not further elevated in fbf-1(ok91) mir-35 fam(0); glp-1(bn18) compared to fbf-1(ok224); glp-1(bn18) (Figure S5C). Therefore, we think it unlikely that the mir-35 family acts to repress GLD-1 in germline stem cells, as previously proposed (Liu et al. 2011). Instead, the mir-35 family may promote a large proliferative zone reminiscent of genes that maintain normal physiology (i.e., eat-2). We did not explore this further.
GLD-1 accumulation pattern is sigmoidal
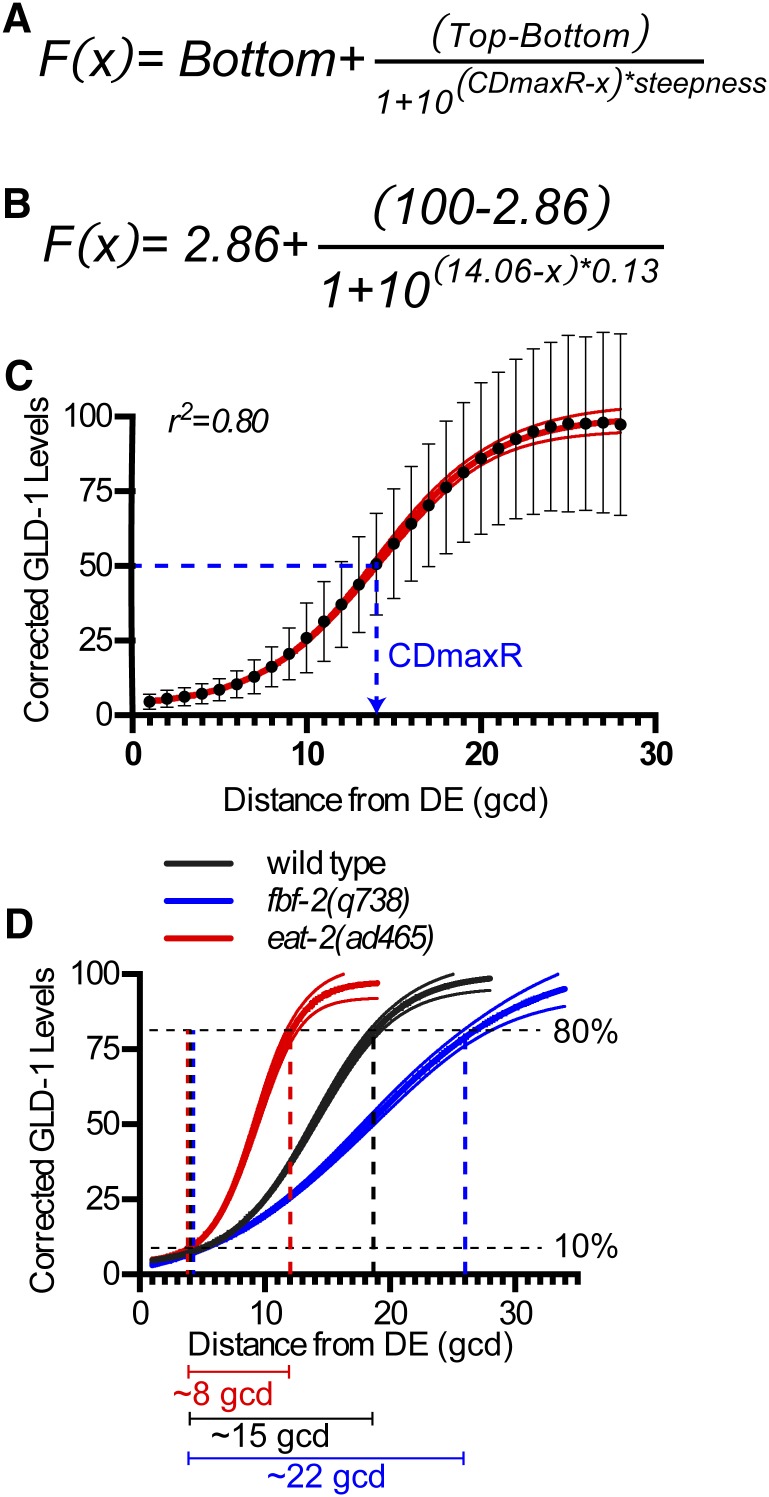
We mathematically modeled GLD-1 levels in the proliferative zone as a means of identifying genetic influences on the accumulation pattern. GLD-1 accumulation appeared sigmoidal and without an obvious linear trend, so we simulated our quantitative data as a dose–response model with four parameters: steepness, CDmaxR, top, and bottom (Figure 7, see Materials and Methods). Steepness is the Hill slope variable in dose–response models and is used as a measure of cooperativity, which we called steepness to reflect our descriptive, rather than functional, interpretation. CDmaxR defines the position, in gcd from distal end, where GLD-1 levels are halfway between top and bottom. We interpreted the CDmaxR value as the position of maximum GLD-1 accumulation change (Figure 7) based on the properties of a symmetrical sigmoidal curve. The bottom and top values are theoretic asymptotic values that were not compared.
Figure 7.
Sigmoidal modeling of GLD-1 accumulation pattern. (A) Modified four-parameter dose–response equation. Bottom and top are not representative of base and peak, but rather theoretical asymptotes used in defining the model. CDmaxR, cell diameter where the ratio of GLD-1 synthesis/GLD-1 turnover is maximum and GLD-1 levels is halfway between top and bottom. (B) Best-fit model equation for wild type. (C) Plot comparing wild-type GLD-1 levels vs. best-fit model. Black dots, mean corrected GLD-1 levels. Red solid line, best-fit model from B. Red dashed lines, 99% confidence interval of best-fit model. r2 = 0.80, weighted correlation. Blue dashed line marks the position of CDmaxR in wild type at ∼14 gcd. (D) Plot comparing best fit-models of genotypes shown, which have the same base and peak but different steepness. Lines are best-fit model ± 99% confidence interval. Bars below graph are span, in gcd, for GLD-1 levels to elevate from 10 to 80% in eat-2 (red), wild type (black), and fbf-2 (blue).
The sigmoidal model correlated with GLD-1 levels in all genotypes analyzed (Figure 7C, Table 3). CDmaxR positively correlated with the sME (Figure S7A), which was expected since the increased GLD-1 accumulation change prefaces overt adoption of meiotic fate. Interestingly, steepness inversely correlated with the sME (Figure S7B). A sigmoidal model also correlated with both GLD-1::GFP accumulation and gld-1 3′ UTR reporter GFP accumulation (Figure S8), supporting that GLD-1 follows a nonlinear, sigmoidal expression pattern and the sigmoidal pattern is not merely a consequence of our assay.
Table 3. Summary of values obtained from best-fit sigmoidal model.
| Genotype | Steepness (95% C.I.)a | CDmaxR (95% C.I.)b | r2c | Comparison of fitd |
|---|---|---|---|---|
| Wild type | 0.13 (0.10–0.14) | 14.1 (13.7–14.4) | 0.80 | N/A |
| gld-1 repression | ||||
| fbf-1(ok91) | 0.12 (0.10–0.14) | 9.6 (9.2–10.1) | 0.80 | ND |
| glp-1(bn18) | 0.17 (0.12–0.21) | 7.1 (6.6–7.5) | 0.78 | ND |
| fbf-1(ok224); glp-1(bn18) | 0.14 (0.09–0.02) | 7.2 (6.1–8.3) | 0.61 | ND |
| fbf-2(q738) | 0.09 (0.08–0.09) | 17.0 (16.3–18.0) | 0.82 | <0.0001 |
| gld-1 activation | ||||
| gld-2(q497) | 0.09 (0.08–0.10) | 14.9 (14.2–15.6) | 0.77 | <0.0001 |
| gld-3(q730) | 0.10 (0.09–0.11) | 11.5 (11.1–12.0) | 0.83 | 0.0028 |
| gld-3(q741) | 0.11 (0.10–0.13) | 13.0 (12.4–13.6) | 0.82 | 0.0565 |
| nos-3(oz231) | 0.14 (0.13–0.16) | 13.6 (13.0–14.2) | 0.80 | 0.0016 |
| Physiological effect | ||||
| eat-2(ad465) | 0.24 (0.22–0.26) | 9.3 (9.0–9.6) | 0.85 | <0.0001 |
| eat-5(ad464) | 0.15 (0.13–0.17) | 13.5 (12.6–14.3) | 0.67 | 0.0017 |
| daf-1(m40) | 0.15 (0.14–0.17) | 11.5 (11.0–12.0) | 0.75 | <0.0001 |
| Putative regulators | ||||
| lst-1(ok814) | 0.11 (0.09–0.14) | 12.0 (11.2–12.8) | 0.78 | <0.0001 |
| sygl-1(tm5040) | 0.24 (0.22–0.27) | 8.9 (8.6–9.2) | 0.82 | <0.0001 |
| mir-35 fam(0)e | 0.14 (0.13–0.16) | 12.2 (11.8–12.6) | 0.79 | 0.0002 |
ND, not determined.
Steepness, analogous to Hill slope value, but changed to reflect its descriptive rather than functional significance for this study.
CDmaxR, position in gcd where GLD-1 level is halfway between top and bottom values (see Figure 7). Interpreted as position where GLD-1 accumulation change is maximum throughout the proliferative zone.
r2 correlation coefficient for best-fit sigmoidal model for a given genotype compared to experimentally determined GLD-1 levels.
Steepness values for each genotype were compared pairwise to the wild-type model by goodness-of-fit test. P < 0.01 considered different. P-value is not calculated when steepness can be described by the same value.
Null(0) for mir-35 family; actual genotype mir-35 thru 41(nDf50) mir-42-45(nDf49).
To illustrate steepness changes, we compared sigmoidal models for wild type, eat-2, and fbf-2 (Figure 7D), since each had similar base and peak GLD-1 levels, but significantly different steepness (Table 3). GLD-1 levels were similar between wild type, eat-2, and fbf-2 in the first ∼4 gcd (Figure 7D), but the distal-to-proximal distance for GLD-1 levels to elevate from 10 to 80% differed (Figure 7D). Genes involved in physiological regulation had elevated steepness (Table 3). Steepness decreased in the absence of genes that promote GLD-1 levels, such as gld-2 and gld-3 (Table 3), suggesting that decreased steepness might serve as a quantifiable characteristic of impaired GLD-1 accumulation across the region where GLD-1 normally accumulates rapidly between base and peak (see Discussion).
GLD-1 levels in germ cells irreversibly committed to meiotic entry
One step in the progression of germline stem cell differentiation is irreversible commitment to the meiotic fate, but where this occurs in the proliferative zone is unknown. Experimental modulation of glp-1 activity through temperature shifting of glp-1(bn18) allows the generation of germ cells identifiable as irreversibly committed (Fox and Schedl 2015) and since high GLD-1 is sufficient to drive meiotic entry (Crittenden et al. 2002; Hansen et al. 2004b), then GLD-1 levels within these germ cells may serve as a protein mark of irreversible commitment in wild type. Removal of glp-1 activity with a 4-hr shift to restrictive temperature, followed by a return to the permissive temperature, results in complete loss of the proliferative zone (Fox and Schedl 2015) (Figure 8). Immediately following the 4-hr pulse at 25°, the distal-most germ cells were REC-8 positive, but by 24 hr after the 4-hr shift, all had entered meiosis (HIM-3 positive) (Figure 8, B and C). Therefore, the REC-8 positive germ cells in glp-1(bn18) after the 4-hr shift to 25° were irreversibly committed to meiosis. In contrast, REC-8 positive germ cells in glp-1(bn18) following a 3-hr shift to 25° were not irreversibly committed to meiosis (Fox and Schedl 2015) (Figure S9). Base GLD-1 levels in glp-1(bn18) adults shifted to 25° for 4 hr were significantly elevated relative to 3 hr (Figure 8F) and were higher than base GLD-1 levels for all other genotypes examined, at ∼40% of peak (Table S2). Base GLD-1 levels showed a minor elevation in wild type when shifted to 25° for 3 or 4 hr (Figure 8F), indicating that the temperature-shift influences GLD-1 levels and levels identified at 25° may not directly scale to levels at 20°. Nonetheless, we extrapolated 40% of peak to wild type at 20° to estimate the position of irreversible commitment at ∼12–13 gcd from the distal end (Figure 8E). This estimated position of irreversible commitment is more proximal than the end of the niche/DTC plexus [∼8–9 gcd (Byrd et al. 2014)] and more distal than the sME [∼19 gcd (Table 1)].
Figure 8.
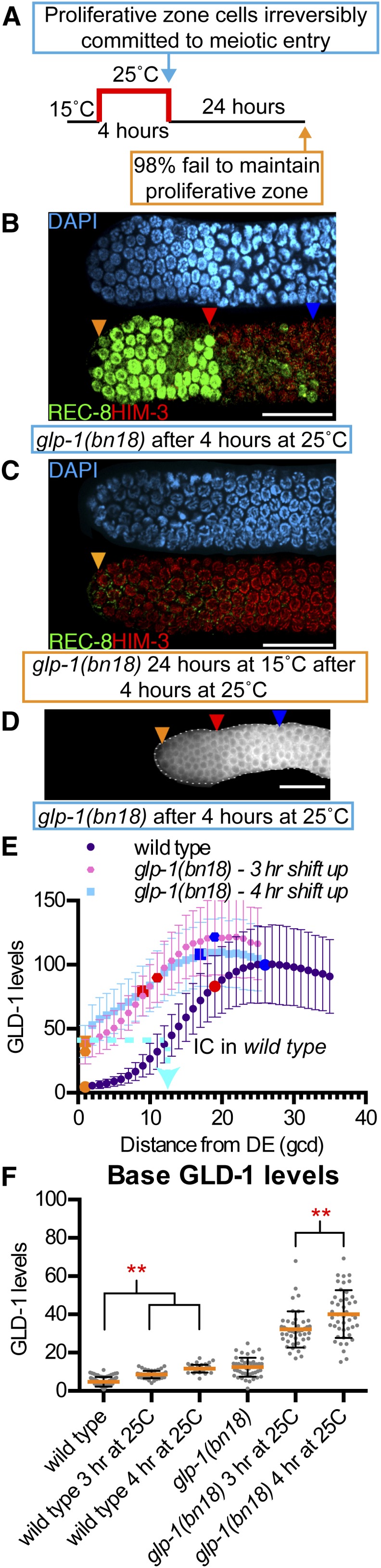
GLD-1 levels in germ cells irreversibly committed to meiotic entry. (A) Growth scheme of glp-1(bn18) to generate proliferative zone cells identifiable as irreversibly committed to meiosis. (B) DAPI and REC-8/HIM-3 immunofluorescence image of glp-1(bn18) after 4 hr at 25°. (C) DAPI and REC-8/HIM-3 immunofluorescence image of glp-1(bn18) 24 hr at 15° after 4 hr at 25°. A total of 41/42 gonads lacked REC-8 positive nuclei as all proliferative zone cells have entered meiosis. (D) Representative GLD-1 immunofluorescence image of glp-1(bn18) after 4 hr at 25°. Orange triangle, 1st gcd; red triangle, sME; blue triangle, eME; white bars in B–D are 25 µm. (E). Plot of GLD-1 levels. Cyan dashed line extends from base GLD-1 level in glp-1(bn18) after 4 hr at 25°, ∼40%, to the same level in wild type, which occurred at ∼12–13 gcd from distal end. We interpreted this as the approximate position of germ cell irreversible commitment (IC) to meiotic development. (F) Scatterplot of GLD-1 base. **P < 0.01, Sidak’s corrected-multiple comparison. Staining results for analysis of glp-1(bn18) shifted for 3 hr at 25° are shown in Figure S9.
Discussion
lst-1 and sygl-1 link glp-1 activity to post-transcriptional control of GLD-1
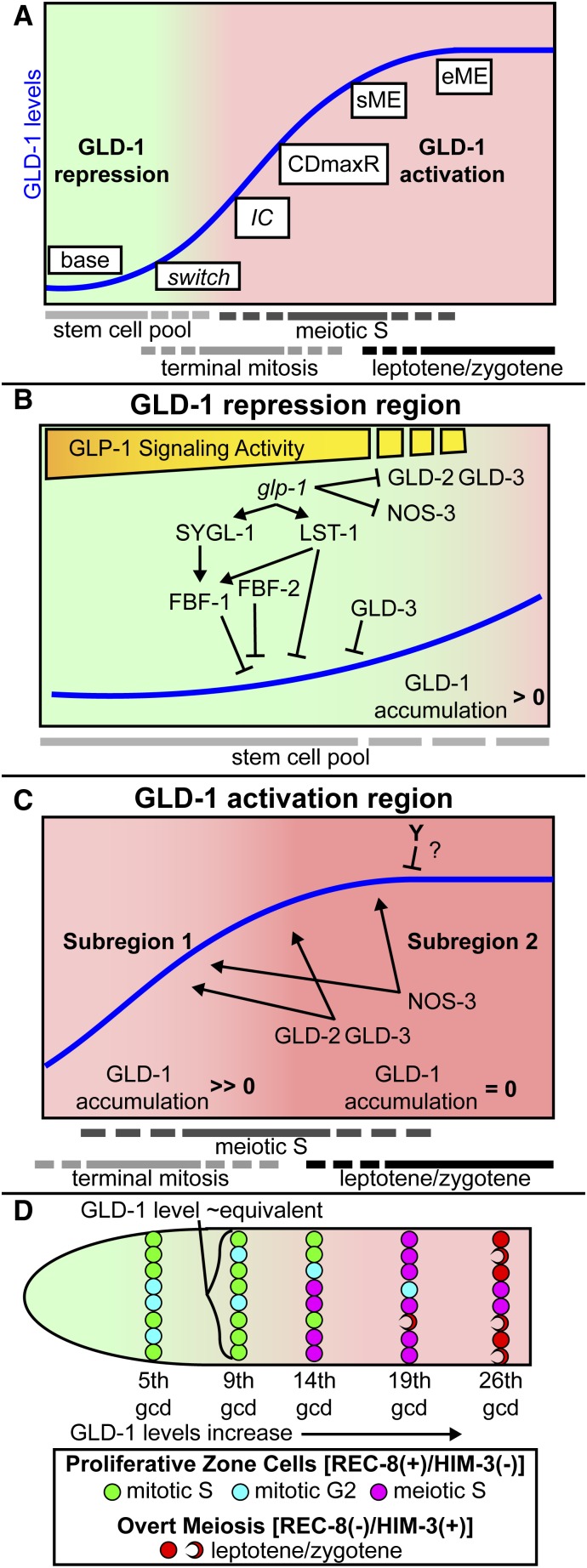
A major gap in our understanding of the proliferative vs. meiotic fate decision was how glp-1-mediated transcription of target genes led to post-transcriptional regulation of GLD-1 accumulation (Crittenden et al. 2002; Hansen et al. 2004b; Suh et al. 2006; Merritt et al. 2008; Suh et al. 2009). We found that sygl-1 and lst-1, direct transcriptional targets of glp-1 signaling (Kershner et al. 2014), act to repress GLD-1 levels in distal germ cells (Figure 5) and therefore place sygl-1 and lst-1 directly downstream of glp-1 in repression of GLD-1 (Figure 9B). sygl-1 likely acts in the same pathway as fbf-1 to repress GLD-1 levels (Figure 6) and since sygl-1 is a direct glp-1 target, we place it upstream to fbf-1 (Figure 9B). lst-1 likely acts in both the same pathway and in parallel to fbf-1 to repress GLD-1 (Figure 6), and since lst-1 is a direct glp-1 target, we place it upstream and in parallel to fbf-1 (Figure 9B). Our data are consistent with a model where GLP-1 signaling controls FBF-1 activity indirectly, via direct transcriptional targets LST-1 and SYGL-1; thus fbf need not be direct transcriptional targets to function downstream of GLP-1 signaling. It is currently unclear if sygl-1 and lst-1 regulate FBF-2. sygl-1 and lst-1 also act in parallel to fbf-1 and fbf-2 to promote germ cell proliferative fate, since sygl-1 and lst-1 each enhanced the premature meiotic entry defects of fbf-1; fbf-2 double mutants (Table 2). Germ cell entry into meiotic prophase requires the redundant GLD-1 and GLD-2 pathways, which are both repressed by glp-1 signaling (Kimble and Crittenden 2007; Hansen and Schedl 2013). Since sygl-1 acts upstream of fbf-1 in GLD-1 repression, but partly in parallel to fbf to promote germ cell proliferative fate, then sygl-1 may also promote proliferative fate in parallel to control of GLD-1 accumulation by repressing the GLD-2 meiotic entry pathway. lst-1 acts in the same pathway and in parallel to fbf-1 to repress GLD-1, and in parallel to fbf-1/-2 to promote germ cell proliferative fate. Therefore, lst-1 may promote germ cell proliferative fate through repression of GLD-1, but we cannot rule out that lst-1 also represses the GLD-2 pathway. FBF also represses synaptonemal complex (SC) genes (Merritt and Seydoux 2010), whose activities are needed during meiotic prophase, and we propose that lst-1 and sygl-1 promote FBF-mediated repression of SC genes similar to their regulation of gld-1.
Figure 9.
Control of GLD-1 accumulation in coordination with germline stem cell differentiation. (A) Model for control of GLD-1 accumulation in the adult hermaphrodite is split into regions called GLD-1 repression and GLD-1 activation. Approximate positions of stem cell pool, progenitors completing their current mitotic cell cycle (terminal mitosis), meiotic S pool, and leptotene/zygotene region are based on previous work (Fox and Schedl 2015). Switch, germ cells exit GLD-1 repression and enter the activation region in coordination with germ cell commitment to meiotic development. IC, irreversible commitment, as experimentally defined by glp-1(bn18) temperature shift, occurs after germ cell commitment and entry into GLD-1 activation. CDmaxR, where rate of GLD-1 accumulation change is maximum closely follows IC as germ cells progress toward sME and eME, where GLD-1 levels plateau. (B) Genetic control of GLD-1 repression region. Germ cells in GLD-1 repression are mostly stem cells, GLD-1 modestly accumulates, and GLD-1 accumulation change is equal or, more proximally, greater than 0. Reduction of GLP-1 signaling activity along the DTC plexus could account for the modestly increased GLD-1 accumulation. (C) Genetic control of GLD-1 activation region is split into two subregions: (1) GLD-1 rapidly accumulates, progenitors complete terminal mitosis and/or initiate meiotic S, and GLD-1 accumulation change is maximum/much greater than 0. (2) GLD-1 levels plateau at peak once all germ cells enter meiosis, and GLD-1 accumulation change = 0. Genes that promote GLD-1 accumulation act in both subregions, but unknown gene(s), Y, downregulates GLD-1 accumulation in subregion 2. Although we measured GLD-1 steady-state levels, we infer that final GLD-1 accumulation output is the ratio of GLD-1 synthesis and GLD-1 turnover, such that when GLD-1 accumulation is >0, then synthesis exceeds turnover, and when GLD-1 accumulation = 0, then GLD-1 synthesis and turnover are equal. (D) GLD-1 levels within germ cells of the same gcd position are similar, even though germ cells are at different stages of the mitotic cell cycle (S or G2), in meiotic S, or in leptotene/zygotene, the proportions of which depend on distance from the distal tip. Germ cells in M phase are low frequency and G1 is short or nonexistent (Fox et al. 2011) therefore not shown.
The molecular activities of SYGL-1 and LST-1 are unknown. sygl-1 encodes a protein that is not obviously conserved outside of nematodes (Kershner et al. 2014). LST-1 contains a Nanos-like zinc finger domain (Kershner et al. 2014), which could confer RNA-binding activity to the gld-1 mRNA, leading to a direct effect on translation efficiency or mRNA stability and/or facilitating binding by FBF. Previous work indicated that an activity separate from FBF-1/-2 represses GLD-1 accumulation in the adult hermaphrodite (Hansen et al. 2004b); lst-1 may provide this parallel activity. Alternatively, LST-1 may promote the activity of another direct repressor of GLD-1 levels. Our data do not support that the mir-35 family are repressors of GLD-1 accumulation, as previously proposed (Liu et al. 2011), and other direct repressors of GLD-1 levels remain to be identified.
Interestingly, we found that gld-3 is required for low GLD-1 base levels, consistent with a role for GLD-3 in repression of GLD-1 levels (Figure 3K). gld-3 is known to promote GLD-1 translation through its interaction with GLD-2 (Eckmann et al. 2004; Suh et al. 2006; Schmid et al. 2009). How gld-3 might act to repress GLD-1 base levels is unclear. GLD-3 may repress GLD-1 indirectly by facilitating binding by direct repressors, such as FBF, based on its proposed function as an RNA–protein scaffold (Nousch and Eckmann 2013).
GLD-1 repression and GLD-1 activation occur regionally
A number of genes that control GLD-1 accumulation are expressed throughout the proliferative zone (Kraemer et al. 1999; Eckmann et al. 2002; Wang et al. 2002; Lamont et al. 2004; Voronina et al. 2012) and we anticipated that these genes would broadly affect GLD-1 levels. Instead, genes involved in repression of GLD-1 only repressed base GLD-1 levels (Figure 2), and genes that activate GLD-1 accumulation promoted high peak GLD-1 levels (Figure 3). The observation that lst-1 and sygl-1 act to repress base may account for the spatial limits of repression and activation, since the mRNA expression of lst-1 and sygl-1 are largely restricted to distal germ cells (Kershner et al. 2014) under glp-1 signaling control. In modeling GLD-1 accumulation, we divide the curve into two regions called GLD-1 repression region and GLD-1 activation region (Figure 9A). Repression occurs from the 1st to ∼7/8th gcd, whereas activation occurs from ∼8/9th to ∼26th gcd (Table S3 and see below). Genes that repress GLD-1 levels function in the GLD-1 repression region, but not measurably in the GLD-1 activation region (Figure 9C). Genes that promote GLD-1 levels function in the GLD-1 activation region, but not measurably in the GLD-1 repression region (Figure 9D).
Regional control of GLD-1 is coordinated with germline stem cell differentiation
In the distal-most germ cells, GLD-1 accumulation is repressed relative to germ cells in more proximal regions. glp-1 signaling and its downstream effectors control GLD-1 accumulation within the GLD-1 repression region (Figure 9B). While low, GLD-1 is detectable in the 1st gcd (Figure S3) and levels increased ∼3- to 4-fold by the 8th gcd (see Table S3), showing a gradual increase in GLD-1 accumulation (Figure 9B), indicating that, although repressed relative to more proximal germ cells in meiotic prophase, germ cells within the GLD-1 repression region have an activating mechanism that permits quantitative expression and a modest increase in GLD-1 levels among the distal-most germ cells. Interestingly, the detectable levels of GLD-1 in distal germ cells do not obviously require the individual activity of genes that promote high peak, such as gld-2 and nos-3, suggesting that (1) their activity may be repressed in germline stem cells (Figure 9B) and (2) their activity alone cannot account for the increase in GLD-1 levels during GLD-1 repression. What activity permits GLD-1 accumulation in the distal-most germ cells is unclear. gld-3 mediates repression within this region by a currently unknown mechanism, so we model it as a repressor separate from the other genes in repression.
GLP-1 signaling strength may diminish as germ cells progress proximally through GLD-1 repression, resulting in reduced repression, or altered modes of repression. FBF-1 and FBF-2 exert different effects on stability and/or localization of gld-1 mRNA (Voronina et al. 2012), and the ratio by which these competing proteins bind to the gld-1 mRNA may be influenced by perceived GLP-1 signaling activity. High levels of LST-1 and/or SYGL-1 may promote FBF-1 binding, thus leading to mRNA destabilization/re-localization and, since mRNA levels are a major contributing factor to protein synthesis, lower GLD-1 levels.
Our analysis suggests there is a change from modest increase (repression) to a rapid increase (activation) in GLD-1 levels. This switch from GLD-1 repression to GLD-1 activation occurs in the vicinity of 7–9 gcd from the distal end, based on the change in GLD-1 level sharply increasing in the proliferative zone at this location (Table S3). Since GLD-1 levels are responsive to GLP-1 signaling activity (Hansen et al. 2004b), then germ cells may relieve GLD-1 repression as they exit the niche/DTC plexus, ∼8–9 gcd from the distal end (Byrd et al. 2014) and simultaneously switch to GLD-1 activation and committing to meiosis. LST-1 and/or SYGL-1 expression, as direct targets of GLP-1 signaling (Kershner et al. 2014), may define the boundary of GLD-1 repression and their downregulation could permit the switch to activation. The expression pattern of LST-1 and SYGL-1 is unknown, but their mRNAs are restricted to distal germ cells within the niche/DTC plexus (Kershner et al. 2014).
We modeled GLD-1 activation into two subregions based on GLD-1 accumulation pattern (Figure 9C). In subregion 1, GLD-1 accumulation change is high. In subregion 2, GLD-1 plateaus at peak level. As germ cells enter subregion 1, they rapidly accumulate GLD-1 to aid in irreversible commitment to meiosis (see below) and to achieve levels necessary for the role of GLD-1 in meiotic prophase. A number of genes are required for rapid accumulation of GLD-1, based on reduced steepness values from sigmoid curve modeling (Table 3). gld-2 and gld-3 regulate GLD-1 accumulation through control of gld-1 mRNA stability and translational efficiency (Suh et al. 2006), and mutants of these genes had small steepness values. The large proliferative zone of fbf-2 mutants is thought to be due to modestly elevated FBF-1 (Lamont et al. 2004; Voronina et al. 2012), which promotes gld-1 mRNA destabilization (Voronina et al. 2012), and may also explain reduced steepness in this mutant. gld-1 mRNA levels may be abnormally low upon the switch to activation in the fbf-2 mutants, which would have an adverse effect on GLD-1 accumulation.
Change in steepness may not be simply related to change in regulation of GLD-1 accumulation. lst-1 had reduced steepness unexpected of its ectopically high GLD-1 levels. nos-3 did not have reduced steepness, despite its role in promoting GLD-1 levels (Hansen et al. 2004b) (Figure 9C). We modeled rapid accumulation of GLD-1 in subregion 1 as requiring gld-2, gld-3, and nos-3 (Figure 9C). Genes that influence physiology, such as eat-2, had increased steepness. However, interpretations of steepness assume equivalent germ cell progression rate and cell cycle dynamics in the proliferative zone between genotypes. The size of the stem cell pool is likely smaller in eat-2 than wild type and, since germ cell progression is driven by cell division, germ cell progression rate from distal to proximal may be slower in eat-2 mutants. Therefore, we did not conclude that genes that influence physiology affect GLD-1 accumulation in subregion 1.
From mathematically modeling GLD-1 accumulation as a sigmoid curve, we identified the CDmaxR position where GLD-1 accumulation change is highest in the proliferative zone (Figure 9C). Immediately after CDmaxR, the rate of GLD-1 accumulation change apparently declines despite the activity of gld-2, gld-3, and nos-3, which are required for the peak in subregion 2, as germ cells begin to overtly adopt the meiotic fate (sME) (Figure 9, A and C). We propose that an unknown activity (Y in Figure 9C) represses GLD-1 levels in subregion 2, but is likely largely absent or inactive during subregion 1 where the apparent rate of GLD-1 accumulation change is high (Figure 9C) and thus explains the plateau of GLD-1 levels at peak when all germ cells have entered meiosis. Presumably plateauing of GLD-1 permits effective downregulation during diplotene/diakinesis (Jones et al. 1996).
Post-translational mechanisms including ubiquitin-mediated proteolysis have been implicated in the control of germline stem cell differentiation (Macdonald et al. 2008; Jeong et al. 2011; Gupta et al. 2015). GLD-1 expression is qualitatively recapitulated by translational reporters (Merritt et al. 2008), and our quantitative analysis of reporter GFP accumulation suggests that most of the quantitative control of GLD-1 levels in the distal germline is mediated through regulation of the gld-1 3′ UTR (Figure S4), indicating that post-translation control of GLD-1 accumulation plays only a minor role in regulating GLD-1 levels in the proliferative zone.
Modeling of germ cell commitment to meiosis through GLD-1 accumulation
The proliferative zone is proposed to be made up of three pools of germ cells that define germline stem cell differentiation: (1) stem cells capable of self-renewing divisions, (2) progenitors undergoing a terminal mitosis, and (3) meiotic S (Fox and Schedl 2015) (Figure 1A, Figure 9). In the proliferative zone, germ cells progress through the cell cycle independently (Fox et al. 2011), such that germ cells in the same distal-to-proximal position are in different phases of the cell cycle and the three pools partially overlap (Figure 9, A and D). We observed that GLD-1 level in an individual cell is most closely associated with its distal–proximal position, not cell cycle stage or point in developmental progression. Nevertheless, we can correlate changes in GLD-1 levels with regulatory changes that are occurring in rows of cells (Figure 9D).
In the distal-most pool (see 5th gcd in Figure 9D), germ cells are undergoing self-renewing mitotic divisions and GLD-1 is repressed. At the 9th gcd, GLD-1 is beginning rapid activation, despite almost all germ cells still being in a mitotic cell cycle. Therefore, progression from stem cell (pool 1) to terminal mitosis (pool 2) likely coincides with germ cells switching from GLD-1 repression to GLD-1 activation. After the 9th gcd, GLD-1 levels elevate rapidly (Table S2) likely to aid in irreversibly committing germ cells to the meiotic fate. We propose germ cells irreversibly commit once GLD-1 reaches a threshold level (Figure 9A, ∼12–13 gcd), based on experimental manipulation of glp-1(bn18) mutants (Figure 8).Many germ cells are still undergoing a mitotic cell cycle upon achieving GLD-1 levels that are correlated with irreversible commitment, but we propose that GLD-1 is at a sufficient level to ensure that the daughters of these mitotic germ cells enter meiotic S. By the 14th gcd, a larger proportion of germ cells are beginning to enter meiotic S, with fewer completing their terminal mitosis (Figure 9D). GLD-1 levels continue to elevate when most germ cells are undergoing meiotic S (19th gcd, Figure 9D), with this elevation presumably needed for meiotic prophase progression/maintenance, and then GLD-1 levels plateau once most/all germ cells have begun leptotene/zygotene (26th gcd, Figure 9D).
Our model of germline stem cell commitment is predicated upon GLD-1 levels and pattern being highly coordinated with germline stem cell differentiation, which is supported by high levels of GLD-1 being sufficient to drive germline stem cell differentiation (Crittenden et al. 2002; Hansen et al. 2004b). However, gld-1 is redundant with the gld-2 pathway in promoting meiotic entry (Kadyk and Kimble 1998; Eckmann et al. 2004; Hansen et al. 2004a) and meiotic entry can occur in the absence of GLD-1, which could compromise GLD-1 levels alone precisely marking commitment to meiosis. We think, in most cases, GLD-1 accumulates in a pattern that accompanies, if not drives, germline stem cell differentiation, since GLD-1 pattern correlated with the position of meiotic entry in all genotypes we analyzed (Figure S6).
Some remaining issues
The polarized pattern of germline stem cell differentiation is initiated by direct glp-1 transcriptional targets, lst-1 and sygl-1, and potentially additional targets, which repress GLD-1 levels. lst-1 and sygl-1 repression is at least in part through promoting fbf-1 activity, but it is unknown if this is through direct interaction with the gld-1 mRNA. It is unclear if lst-1 and sygl-1 also promote fbf-2 and thus limit its activity to the stem cell region. How gld-2 and nos-3 activity is apparently blocked during GLD-1 repression is not well defined, but may also be through glp-1 signaling. The switch from repression to activation presumably involves spatial restriction of lst-1 and sygl-1 to distal-most germ cells. However, additional gene activities are likely required for the remodeling/relocalization of the gld-1 mRNA necessary to convert from repression to active translation. At the mechanistic level, the contribution of translational vs. mRNA stability control for each regulator remains to be determined. Nonoptimal physiological conditions shrink the proliferative zone compressing the GLD-1 accumulation curve without affecting base or peak GLD-1 levels; how this more subtle modification in GLD-1 accumulation occurs is unknown.
Acknowledgments
We thank the members of the Schedl, Kornfeld, Nonet, and Pincus labs for helpful discussions. Antibodies for REC-8 were provided by Verena Jantsch and Joseph Loidl, HIM-3 by Monique Zetka, and GFP by Swathi Arur. This work is supported by National Institutes of Health (NIH) F32GM106615 to J.L.B. and R01GM100756 to T.S. Some strains were provided by the Caenorhabditis Genetics Center, which is funded by NIH Office of Research Infrastructure Programs (P40-OD010440).
Footnotes
Communicating editor: D. I. Greenstein
Supporting information is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.185678/-/DC1.
Literature Cited
- Andersson E. R., Sandberg R., Lendahl U., 2011. Notch signaling: simplicity in design, versatility in function. Development 138: 3593–3612. [DOI] [PubMed] [Google Scholar]
- Austin J., Kimble J., 1987. glp-1 is required in the germ line for regulation of the decision between mitosis and meiosis in C. elegans. Cell 51: 589–599. [DOI] [PubMed] [Google Scholar]
- Avery L., 1993. The genetics of feeding in Caenorhabditis elegans. Genetics 133: 897–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biedermann B., Wright J., Senften M., Kalchhauser I., Sarathy G., et al. , 2009. Translational repression of cyclin E prevents precocious mitosis and embryonic gene activation during C. elegans meiosis. Dev. Cell 17: 355–364. [DOI] [PubMed] [Google Scholar]
- Bray S. J., 2006. Notch signalling: a simple pathway becomes complex. Nat. Rev. Mol. Cell Biol. 7: 678–689. [DOI] [PubMed] [Google Scholar]
- Byrd D. T., Kimble J., 2009. Scratching the niche that controls Caenorhabditis elegans germline stem cells. Semin. Cell Dev. Biol. 20: 1107–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd D. T., Knobel K., Affeldt K., Crittenden S. L., Kimble J., 2014. A DTC niche plexus surrounds the germline stem cell pool in Caenorhabditis elegans. PLoS One 9: e88372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciosk R., DePalma M., Priess J. R., 2006. Translational regulators maintain totipotency in the Caenorhabditis elegans germline. Science 311: 851–853. [DOI] [PubMed] [Google Scholar]
- Crittenden S. L., Bernstein D. S., Bachorik J. L., Thompson B. E., Gallegos M., et al. , 2002. A conserved RNA-binding protein controls germline stem cells in Caenorhabditis elegans. Nature 417: 660–663. [DOI] [PubMed] [Google Scholar]
- Dalfó D., Michaelson D., Hubbard E. J. A., 2012. Sensory regulation of the C. elegans germline through TGF-β-dependent signaling in the niche. Curr. Biol. 22: 712–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dernburg A. F., McDonald K., Moulder G., Barstead R., Dresser M., et al. , 1998. Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell 94: 387–398. [DOI] [PubMed] [Google Scholar]
- Eckmann C. R., Kraemer B., Wickens M., Kimble J., 2002. GLD-3, a bicaudal-C homolog that inhibits FBF to control germline sex determination in C. elegans. Dev. Cell 3: 697–710. [DOI] [PubMed] [Google Scholar]
- Eckmann C. R., Crittenden S. L., Suh N., Kimble J., 2004. GLD-3 and control of the mitosis/meiosis decision in the germline of Caenorhabditis elegans. Genetics 168: 147–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox P. M., Schedl T., 2015. Analysis of germline stem cell differentiation following loss of GLP-1 Notch activity in Caenorhabditis elegans. Genetics 201: 167–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox P. M., Vought V. E., Hanazawa M., Lee M.-H., Maine E. M., et al. , 2011. Cyclin E and CDK-2 regulate proliferative cell fate and cell cycle progression in the C. elegans germline. Development 138: 2223–2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis R., Barton M. K., Kimble J., Schedl T., 1995a gld-1, a tumor suppressor gene required for oocyte development in Caenorhabditis elegans. Genetics 139: 579–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis R., Maine E., Schedl T., 1995b Analysis of the multiple roles of gld-1 in germline development: interactions with the sex determination cascade and the glp-1 signaling pathway. Genetics 139: 607–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgi L. L., Albert P. S., Riddle D. L., 1990. daf-1, a C. elegans gene controlling dauer larva development, encodes a novel receptor protein kinase. Cell 61: 635–645. [DOI] [PubMed] [Google Scholar]
- Gupta P., Leahul L., Wang X., Wang C., Bakos B., et al. , 2015. Proteasome regulation of the chromodomain protein MRG-1 controls the balance between proliferative fate and differentiation in the C. elegans germ line. Development 142: 291–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen D., Schedl T., 2013. Stem cell proliferation vs. meiotic fate decision in Caenorhabditis elegans. Adv. Exp. Med. Biol. 757: 71–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen D., Hubbard E. J. A., Schedl T., 2004a Multi-pathway control of the proliferation vs. meiotic development decision in the Caenorhabditis elegans germline. Dev. Biol. 268: 342–357. [DOI] [PubMed] [Google Scholar]
- Hansen D., Wilson-Berry L., Dang T., Schedl T., 2004b Control of the proliferation vs. meiotic development decision in the C. elegans germline through regulation of GLD-1 protein accumulation. Development 131: 93–104. [DOI] [PubMed] [Google Scholar]
- Henderson S. T., Gao D., Lambie E. J., Kimble J., 1994. lag-2 may encode a signaling ligand for the GLP-1 and LIN-12 receptors of C. elegans. Development 120: 2913–2924. [DOI] [PubMed] [Google Scholar]
- Hubbard E. J. A., Korta D. Z., Dalfó D., 2013. Physiological control of germline development. Adv. Exp. Med. Biol. 757: 101–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J., Verheyden J. M., Kimble J., 2011. Cyclin E and Cdk2 control GLD-1, the mitosis/meiosis decision, and germline stem cells in Caenorhabditis elegans. PLoS Genet. 7: e1001348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones A. R., Francis R., Schedl T., 1996. GLD-1, a cytoplasmic protein essential for oocyte differentiation, shows stage- and sex-specific expression during Caenorhabditis elegans germline development. Dev. Biol. 180: 165–183. [DOI] [PubMed] [Google Scholar]
- Jungkamp A.-C., Stoeckius M., Mecenas D., Grün D., Mastrobuoni G., et al. , 2011. In vivo and transcriptome-wide identification of RNA binding protein target sites. Mol. Cell 44: 828–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadyk L. C., Kimble J., 1998. Genetic regulation of entry into meiosis in Caenorhabditis elegans. Development 125: 1803–1813. [DOI] [PubMed] [Google Scholar]
- Kershner A. M., Kimble J., 2010. Genome-wide analysis of mRNA targets for Caenorhabditis elegans FBF, a conserved stem cell regulator. Proc. Natl. Acad. Sci. USA 107: 3936–3941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kershner A. M., Shin H., Hansen T. J., Kimble J., 2014. Discovery of two GLP-1/Notch target genes that account for the role of GLP-1/Notch signaling in stem cell maintenance. Proc. Natl. Acad. Sci. USA 111: 3739–3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. W., Wilson T. L., Kimble J., 2010. GLD-2/RNP-8 cytoplasmic poly(A) polymerase is a broad-spectrum regulator of the oogenesis program. Proc. Natl. Acad. Sci. USA 107: 17445–17450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimble J., Crittenden S. L., 2007. Controls of germline stem cells, entry into meiosis, and the sperm/oocyte decision in Caenorhabditis elegans. Annu. Rev. Cell Dev. Biol. 23: 405–433. [DOI] [PubMed] [Google Scholar]
- Kimble J. E., White J. G., 1981. On the control of germ cell development in Caenorhabditis elegans. Dev. Biol. 81: 208–219. [DOI] [PubMed] [Google Scholar]
- Kodoyianni V., Maine E. M., Kimble J., 1992. Molecular basis of loss-of-function mutations in the glp-1 gene of Caenorhabditis elegans. Mol. Biol. Cell 3: 1199–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korta D. Z., Tuck S., Hubbard E. J. A., 2012. S6K links cell fate, cell cycle and nutrient response in C. elegans germline stem/progenitor cells. Development 139: 859–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer B., Crittenden S., Gallegos M., Moulder G., Barstead R., et al. , 1999. NANOS-3 and FBF proteins physically interact to control the sperm-oocyte switch in Caenorhabditis elegans. Curr. Biol. 9: 1009–1018. [DOI] [PubMed] [Google Scholar]
- Lamont L. B., Crittenden S. L., Bernstein D., Wickens M., Kimble J., 2004. FBF-1 and FBF-2 regulate the size of the mitotic region in the C. elegans germline. Dev. Cell 7: 697–707. [DOI] [PubMed] [Google Scholar]
- Liu J., Sato C., Cerletti M., Wagers A., 2010. Notch signaling in the regulation of stem cell self-renewal and differentiation. Curr. Top. Dev. Biol. 92: 367–409. [DOI] [PubMed] [Google Scholar]
- Liu M., Liu P., Zhang L., Cai Q., Gao G., et al. , 2011. mir-35 is involved in intestine cell G1/S transition and germ cell proliferation in C. elegans. Cell Res. 21: 1605–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald L. D., Knox A., Hansen D., 2008. Proteasomal regulation of the proliferation vs. meiotic entry decision in the Caenorhabditis elegans germ line. Genetics 180: 905–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt C., Seydoux G., 2010. The Puf RNA-binding proteins FBF-1 and FBF-2 inhibit the expression of synaptonemal complex proteins in germline stem cells. Development 137: 1787–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt C., Rasoloson D., Ko D., Seydoux G., 2008. 3′ UTRs are the primary regulators of gene expression in the C. elegans germline. Curr. Biol. 18: 1476–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelson D., Korta D. Z., Capua Y., Hubbard E. J. A., 2010. Insulin signaling promotes germline proliferation in C. elegans. Development 137: 671–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadarajan S., Govindan J. A., McGovern M., Hubbard E. J. A., Greenstein D., 2009. MSP and GLP-1/Notch signaling coordinately regulate actomyosin-dependent cytoplasmic streaming and oocyte growth in C. elegans. Development 136: 2223–2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nousch M., Eckmann C. R., 2013. Translational control in the Caenorhabditis elegans germ line. Adv. Exp. Med. Biol. 757: 205–247. [DOI] [PubMed] [Google Scholar]
- Nousch M., Yeroslaviz A., Habermann B., Eckmann C. R., 2014. The cytoplasmic poly(A) polymerases GLD-2 and GLD-4 promote general gene expression via distinct mechanisms. Nucleic Acids Res. 126: 4274–4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasierbek P., Jantsch M., Melcher M., Schleiffer A., Schweizer D., et al. , 2001. A Caenorhabditis elegans cohesion protein with functions in meiotic chromosome pairing and disjunction. Genes Dev. 15: 1349–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M., Küchler B., Eckmann C. R., 2009. Two conserved regulatory cytoplasmic poly(A) polymerases, GLD-4 and GLD-2, regulate meiotic progression in C. elegans. Genes Dev. 23: 824–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher B., Hanazawa M., Lee M.-H., Nayak S., Volkmann K., et al. , 2005. Translational repression of C. elegans p53 by GLD-1 regulates DNA damage-induced apoptosis. Cell 120: 357–368. [DOI] [PubMed] [Google Scholar]
- Suh N., Jedamzik B., Eckmann C. R., Wickens M., Kimble J., 2006. The GLD-2 poly(A) polymerase activates gld-1 mRNA in the Caenorhabditis elegans germ line. Proc. Natl. Acad. Sci. USA 103: 15108–15112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh N., Crittenden S. L., Goldstrohm A., Hook B., Thompson B., et al. , 2009. FBF and its dual control of gld-1 expression in the Caenorhabditis elegans germline. Genetics 181: 1249–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tax F. E., Yeargers J. J., Thomas J. H., 1994. Sequence of C. elegans lag-2 reveals a cell-signalling domain shared with Delta and Serrate of Drosophila. Nature 368: 150–154. [DOI] [PubMed] [Google Scholar]
- Voronina E., Paix A., Seydoux G., 2012. The P granule component PGL-1 promotes the localization and silencing activity of the PUF protein FBF-2 in germline stem cells. Development 139: 3732–3740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Eckmann C. R., Kadyk L. C., Wickens M., Kimble J., 2002. A regulatory cytoplasmic poly(A) polymerase in Caenorhabditis elegans. Nature 419: 312–316. [DOI] [PubMed] [Google Scholar]
- Waters J. C., 2009. Accuracy and precision in quantitative fluorescence microscopy. J. Cell Biol. 185: 1135–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf D. E., Samarasekera C., Swedlow J. R., 2013. Quantitative analysis of digital microscope images. Methods Cell Biol. 114: 337–367. [DOI] [PubMed] [Google Scholar]
- Wood W. B., 1988. The Nematode Caenorhabditis elegans, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Zetka M. C., Kawasaki I., Strome S., Müller F., 1999. Synapsis and chiasma formation in Caenorhabditis elegans require HIM-3, a meiotic chromosome core component that functions in chromosome segregation. Genes Dev. 13: 2258–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Gallegos M., Puoti A., Durkin E., Fields S., et al. , 1997. A conserved RNA-binding protein that regulates sexual fates in the C. elegans hermaphrodite germ line. Nature 390: 477–484. [DOI] [PubMed] [Google Scholar]