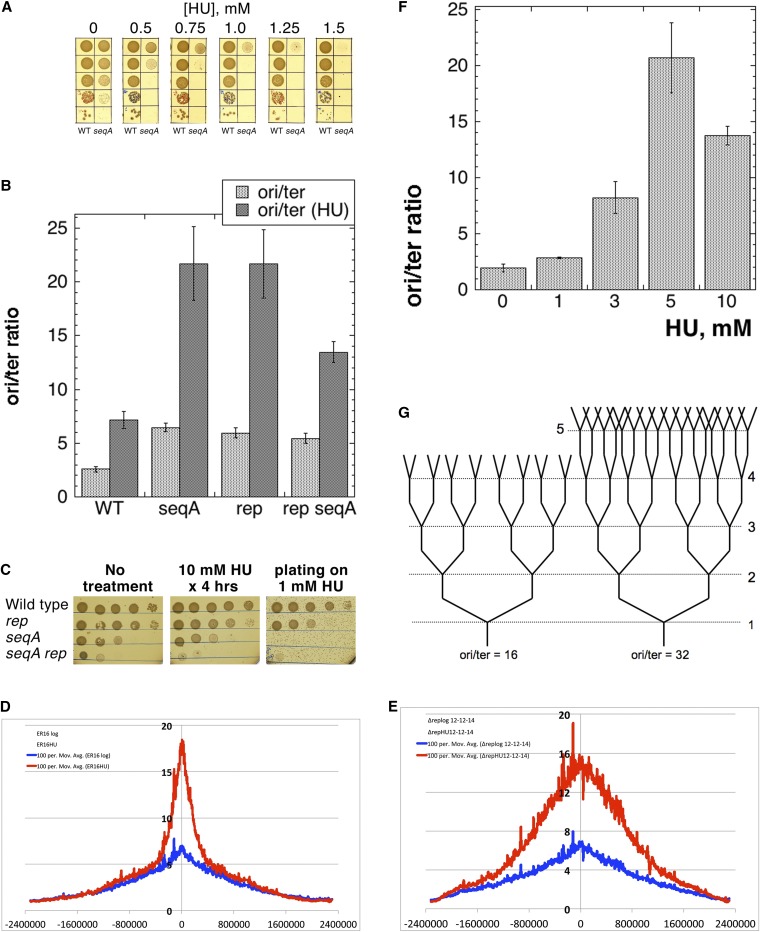
Figure 2.
Breaking through the natural limit of chromosomal replication complexity reveals the “functional” CRC limit. (A) Inhibition of seqA mutant cells with concentrations of HU subinhibitory for wild-type cells. (B) Replication complexity in seqA, rep, and seqA rep mutant cells treated with 10 mM HU for 4 hr. Data for wild-type (HU) and for untreated mutants are from Figure 1B. (C) Survival of the cells from cultures either treated with 10 mM HU for 4 hr or spotted on LB + 1 mM HU. (D) Chromosomal marker-frequency profiles of the untreated seqA mutant (blue trend line) vs. the seqA mutant treated with 10 mM HU for 4 hr (red trend line). (E) Chromosomal marker-frequency profiles of the untreated rep mutant (blue trend line) vs. the rep mutant treated with 10 mM HU for 4 hr (red trend line). (F) CRC in wild-type cells growing at steady state in the presence of the indicated concentrations of HU. An overnight culture of AB1157 was diluted 1000-fold and split into several subcultures grown in the presence of the indicated HU concentrations for 24 hr. Subcultures that reached saturation were again diluted 100-fold into the same media and grown for ∼4 hr. The 10 mM culture grew extremely slowly and was not diluted the second time, but just grown for another 4 hr. Kinetics of OD increase of the cultures were monitored, as well as the shape of the cells under the microscope, to guard against a sweep by HU-resistant mutants. (G) Schematic chromosomal replication complexity at ori/ter = 16 and 32.