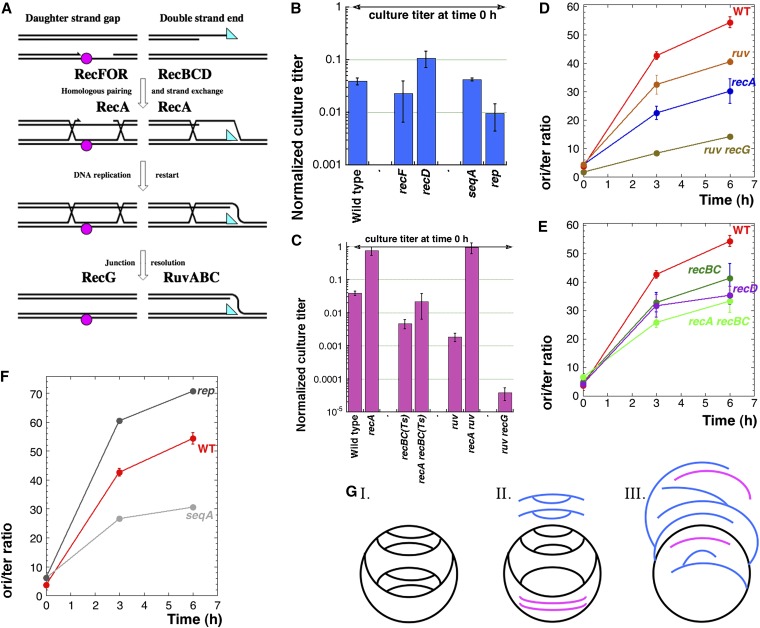
Figure 5.
Genetics of survival of the induced overinitiation vs. the kinetics of CRC accumulation. (A) A scheme of the two major pathways of recombinational repair in E. coli. Magenta circle, a noncoding DNA lesion; cyan triangle, the invading 3′-single-strand end. (B) Survival of 6 hr IPTG induction by wild-type and various mutant IOC cultures with wild-type-like effects. (C) Survival of 6 hr IPTG induction by wild-type and various double-strand break repair-mutant IOC cultures. (D) Kinetics of the ori/ter ratios during IPTG induction of IOC in wild-type cells, as well as in recA, ruv, and ruv recG mutants. (E) Kinetics of the ori/ter ratios during IPTG induction of IOC in recBC(Ts), recA recBC(Ts), and recD mutants. (F) Kinetics of the ori/ter ratios during IPTG induction of IOC in rep and seqA mutants. (G) A scheme of possible chromosome problems due to runaway CRC. DNA duplexes are presented by single lines. Black lines, the circular domains of the chromosome; blue lines, the (partially) linear domains that would not enter pulsed-field gels; the pink lines, subchromosomal fragments that would enter pulsed-field gels. Scenario I: The overreplicating chromosome without problems and lethality. Scenario II: Some replication forks rear-ended the previous forks in a paired fashion, while (in the top half) new initiations also happened, masking some fragmentation. Similar to scenario I, this scenario also predicts no lethality, even without any further action form the cell. Scenario III: Unpaired replication fork disintegration generates multi-tailed chromosome (expected to be lethal without repair or linear DNA degradation).