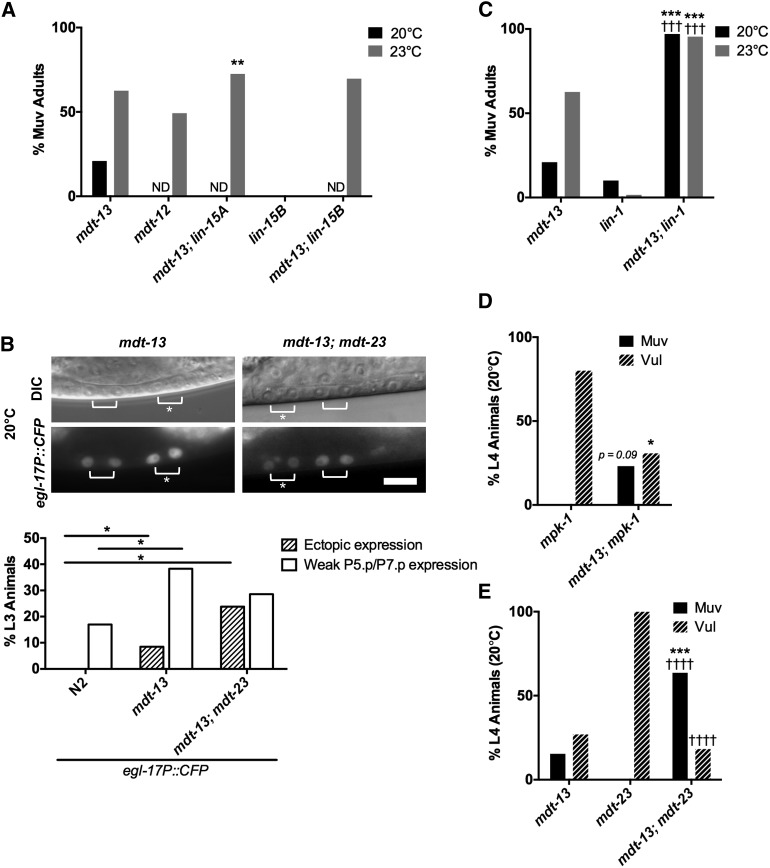
Figure 5.
mdt-13/let-19 represses vulval cell fates downstream of mpk-1/ERK. (A) Adult Muv penetrance in mdt-13/let-19 mutants and mutants with representative synMuv genes (n ≥ 99). **P < 0.01 vs. mdt-13/let-19 single mutant, Fisher’s exact test. ND, not determined. See Table S2 for raw data. (B) Micrographs show mdt-13/let-19 and mdt-13/let-19; mdt-23/sur-2 mutants expressing the arIs92[egl-17P::CFP] 1° fate marker. Bracket: Pn.px cells expressing reporter. Asterisk: ectopic reporter expression. Bar: 10 μm. Graphs show the percentage of animals (n > 20) with ectopic egl-17P::CFP expression (defined as expression in P3.p/P4.p/P8.p, or expression in P5.p/P7.p of equal intensity to P6.p) or weak P5.p/P7.p expression (defined as expression in P5.p/P7.p that is weaker intensity than P6.p). *P < 0.05, Fisher’s exact test. (C) Adult Muv penetrance in mdt-13/let-19; lin-1(n1790) mutants (n ≥ 44). mdt-13/let-19 single mutant Muv penetrance from Figure 5A is included for reference. ***P < 0.001 vs. mdt-13/let-19 single mutant, †††P < 0.001 vs. lin-1(n1790) single mutant, Fisher’s exact test. See Table S2 for raw data. (D) L4 Muv and Vul penetrance in mdt-13/let-19; mpk-1/ERK mutants (n ≥ 13). P = 0.09 (Muv) and *P < 0.05 (Vul) vs. mpk-1 single mutant, Fisher’s exact test. See Table 1 for raw data. (E) L4 Muv and Vul penetrance in mdt-13/let-19; mdt-23/sur-2 mutants (n ≥ 22). ***P < 0.001 vs. mdt-13/let-19 single mutant, ††††P < 0.0001 vs. mdt-23/sur-2 single mutant, Fisher’s exact test. See Table 1 for raw data.