Figure 4.
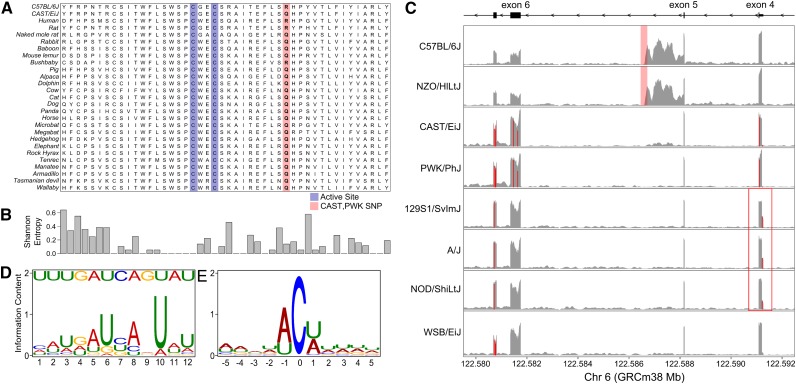
Genomic variants in the DO founders fit the pattern of Apob editing. (A) A nonsynonymous SNP in exon 6 of Apobec1 contributed by CAST/EiJ and PWK/PhJ converts an arginine (R) residue to a glutamine (Q), highlighted in red. Residues in the active site are highlighted in blue. (B) Most mammals have a glutamine residue at this location. Shannon entropy for each base position shows that the glutamine is somewhat conserved. (C) RNA-seq pileups of Apobec1 in the eight DO founders show transcriptional variation between alleles. C57BL/6J and NZO/HlLtJ alleles carry an insertion (shaded in red) in the fifth intron that overlaps a retained intron. 129S1/SvImJ, A/J, and NOD/ShiLtJ alleles carry a SNP (red box) in the 5′ UTR of exon 4 that has increased expression in those strains. The y-axis shows the read depth normalized to library size. (D) Consensus mooring sequence motif for C-to-U edited genes. (Top) The Apobec1 mooring sequence. (Bottom) The consensus binding motif discovered using MEME. (E) Sequence information content around the edited C-to-U site shows that bases at the proximal and distal positions are either A or U. There does not appear to be sequence preference at other nearby base positions.