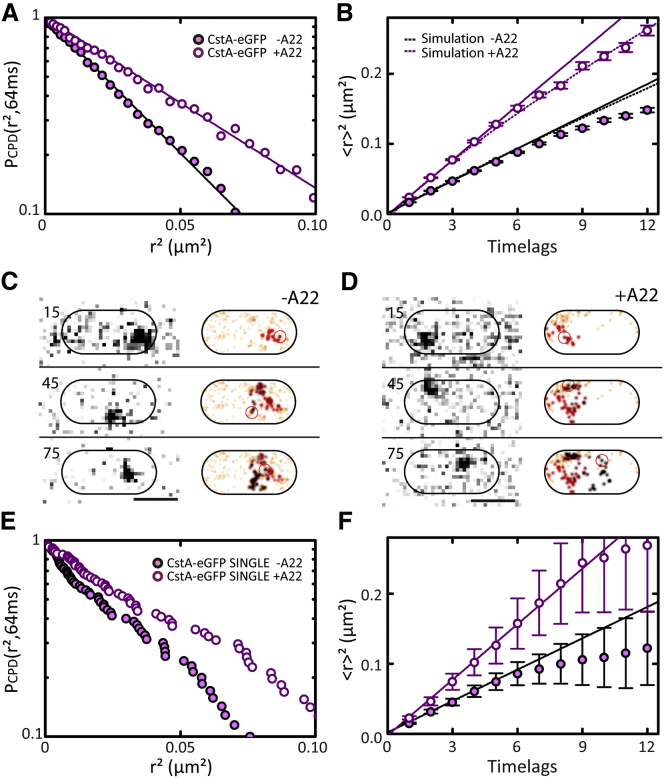
Figure 4.
TMP diffusion in the presence and absence of polymerized MreB. (A) CPD analysis of CstA-eGFP diffusion in the absence (solid symbols) and presence (open symbols) of MreB-polymerization inhibitor A22. (Solid lines) Fits with single-exponential functions (Table S2). (B) MSD plots of experimental data shown in (A). (Solid lines) Linear fits on the first four time lags (Table 1). (Dotted lines) Nonlinear fits on MSD values simulated by a random walk with the same diffusion coefficient over the part of a bacterial cell that is observable by wide-field epifluorescence microscopy. (C and D) Individual, long single-molecule trajectory of CstA-eGFP in absence (C) and presence (D) of A22. (Left panel) Sequence of raw images; (right panel) 2D-Gaussian-rendered single-molecule positions of the trajectory, color-coded according to their occurrence in time from red to black. (Orange) Pre- and succeeding single-molecule events recorded within the observed bacterium. Frame numbers are indicated on the left; time interval between two consecutive frames = 32 ms. (E and F) CPD and MSD analysis, respectively, on the individual single-molecule trajectories of CstA-eGFP from (C) and (D). (F) Fits were performed as in (B). Color-coding as in (A).