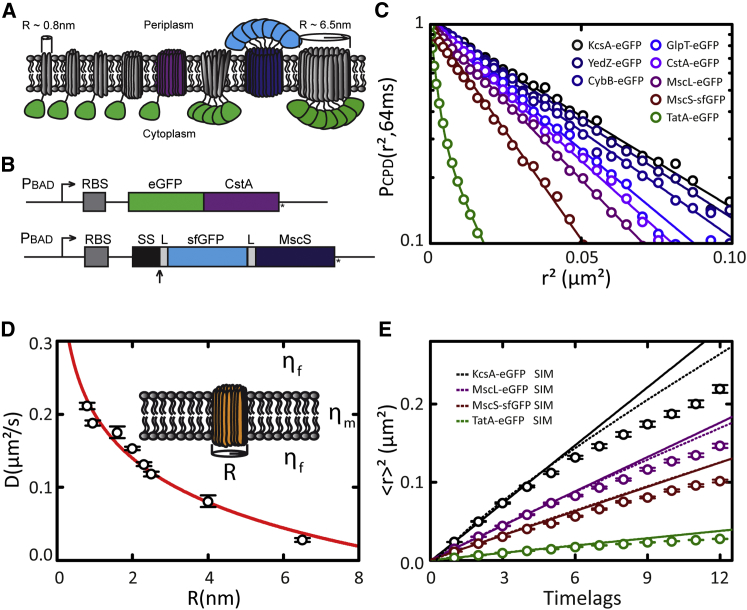
Figure 5.
Size-dependent TMP diffusion in the cytoplasmic membrane of E. coli bacteria. (A) Illustration of the library of the eight GFP-tagged TMPs used in this study (see Table 1 for names and radii). (B) Schematic of GFP fusion constructs: (top) eGFP (green) fused to the amino-terminus of CstA. (Bottom) sfGFP (light blue) fused to the periplasmic amino-terminus of MscS, preceded by the signal sequence (SS) of DsbA, to achieve cotranslational translocation of sfGFP. (Arrow) Site of leader peptidase processing; L indicates a short flexible linker. (C) CPD analysis of TMP diffusion within time lag of 64 ms. (Straight, solid lines) Single-exponential fits to the CPD data. In the case of TatA-eGFP, a double-exponential function was fitted. Fit results are presented in Table S2. (D) Diffusion coefficients obtained using MSD analysis (Table 1) plotted against radius R of corresponding TMP. Fitting the Saffman-Delbrück model yields a membrane viscosity μm of 1.2 ± 0.1 Pa·s and bulk viscosity μf of 0.24 ± 0.02 Pa∙s. (E) MSD analysis of KcsA-eGFP, MscL-eGFP, MscS-sfGFP, and TatA-eGFP. Time lag = 32 ms. (Solid lines) Linear fits on the first four time lags (Table 1). (Dotted lines) Nonlinear fits on MSD values simulated by a random walk with the same diffusion coefficient over the part of a bacterial cell that is observable by wide-field epifluorescence microscopy. Color-coding as in (C).