Figure 2.
Effects of 2HG on Histone Demethylation, Activation of Pluripotency Genes, and Genome-wide DNA Methylation
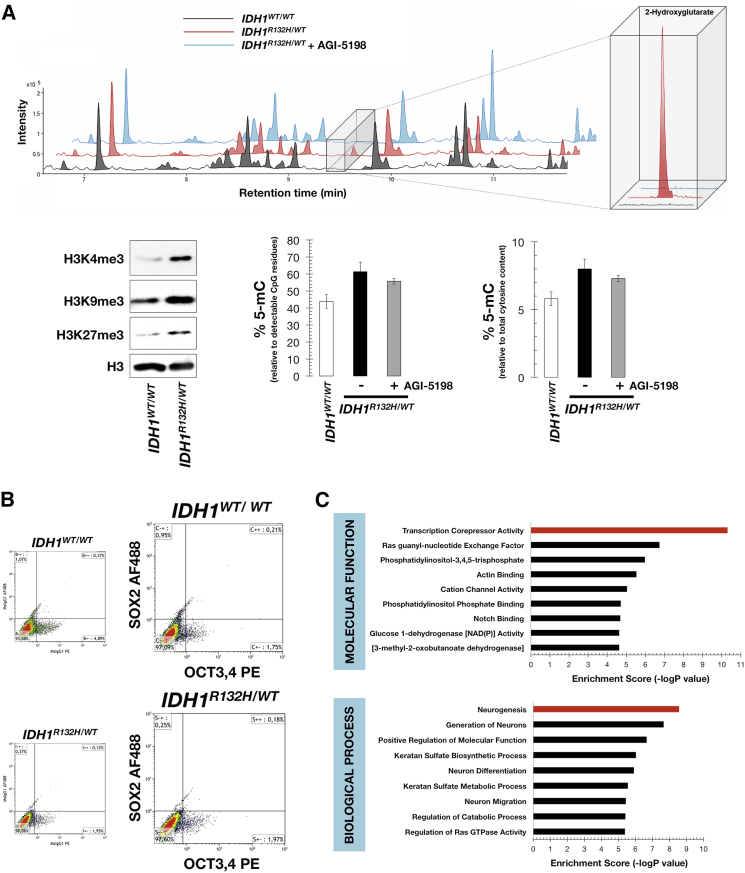
(A) Top: Base-peak chromatograms of extracts from IDH1WT/WT cells (black line), IDH1R132H/WT cells (red line), and IDH1R132H/WT cells treated with 40 μmol/l AGI-5198 for 2 days (blue line). A combined mass spectrum of the region where 2HG was eluted (m/z 349.1317) is shown in the inset (two technical replicates per n; n = 3 biological replicates). Bottom: 2HG promotes broad inhibition of histone demethylation and LINE-1 global methylation. Left panel, western blots for total H3K4me3, H3K9me3, and H3K27me3 histone modifications in parental IDH1WT/WT and IDH1R132H/WT knockin cells. Also shown are total H3 controls (two technical replicates per n; n = 2 biological replicates). Middle and right panels, % of 5-mC relative to either detectable CpG residues or total cytosine content in IDH1WT/WT and IDH1R132H/WT cells, the latter being cultured in the absence or presence of 40 μmol/l AGI-5198 for 2 days. The data are presented as the mean ± SD (error bars); three technical replicates per n; n = 2 biological replicates.
(B and C) 2HG impairs differentiation but does not elevate the baseline expression of core pluripotency factors. (B) Flow cytometry analysis of OCT4/SOX2 expression in IDH1WT/WT and IDH1R132H/WT knockin cells. Representative dot plots showing the distribution of IDH1WT/WT and IDH1R132H/WT cells along the signal obtained with the isotype-specific control antibodies or with the OCT3,4 and SOX2 direct conjugated antibodies (two technical replicates per n; n = 2 biological replicates). (C) Ten top-ranked GO molecular functions and biological processes associated with hypermethylated genes in IDH1R132H/WT cells (accession number GEO: GSE76263). x axis, negative logarithm (-lg) of the p value; y axis, GO category.