Figure 4.
Oncometabolite-Driven Nuclear Reprogramming of Cancer Stemness: A Framework Proposal
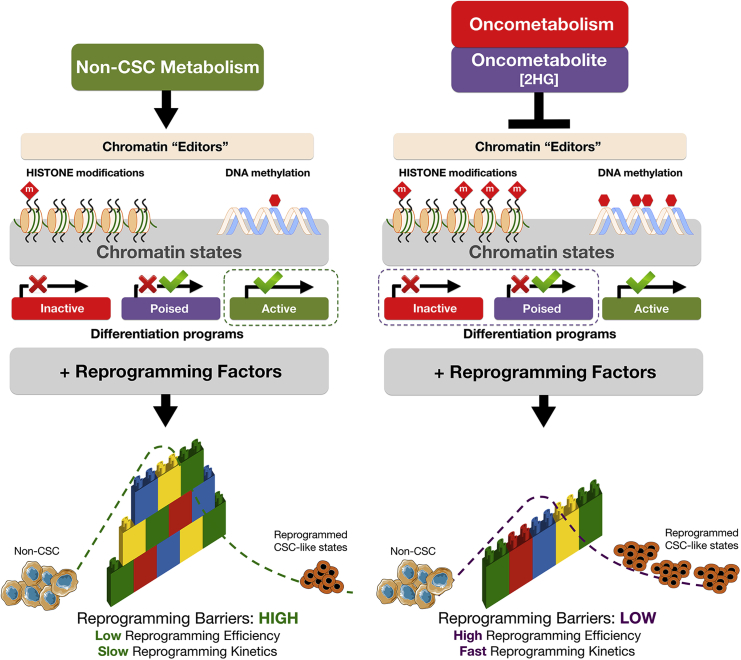
HDMs, such as Jumonji histone demethylases (JHDM) and ten-eleven translocation (TET) family members, remove repressive histone methylation marks and activate the expression of differentiation-related genes by protecting promoters from aberrant DNA methylation. Oncometabolites such as 2HG inhibit the epigenetic “editors” HDMs and TETs, which leads to histone modifications (e.g., increased H3K9me3, H3K27me3, and H3K4me3) and DNA hypermethylation. Oncometabolites reprogram chromatin state to promote the downregulation of genes involved in differentiation as well as bias in developmental gene-expression patterns. This metabolo-epigenetic modification of inactive/poised states of lineage-specific genes is sufficient to significantly alter the efficiency and speed of nuclear reprogramming by lowering the “reprogramming barriers” of the epigenetic landscape and increasing the size of the stem cell state basin of attraction, which results in the acceleration (i.e., higher efficiency and faster kinetics) of the nuclear reprogramming process. Oncometabolites such as 2HG permissively alleviate the unfavorable developmental process of “jumping” from differentiated cell states to CSC-like attractors while concomitantly stabilizing the ground-state self-maintaining character of CSC states. This conceptual figure represents cells stabilized in an initial non-CSC attractor and how nuclear reprogramming can make cells exceed the “reprogramming barrier,” represented as a wall of interlocking bricks, harder or easier in the absence or presence of the oncometabolite 2HG, respectively, and fall down in a final CSC attractor. The cellular reprogramming process is represented as a dashed line from the initial to the final cellular state.