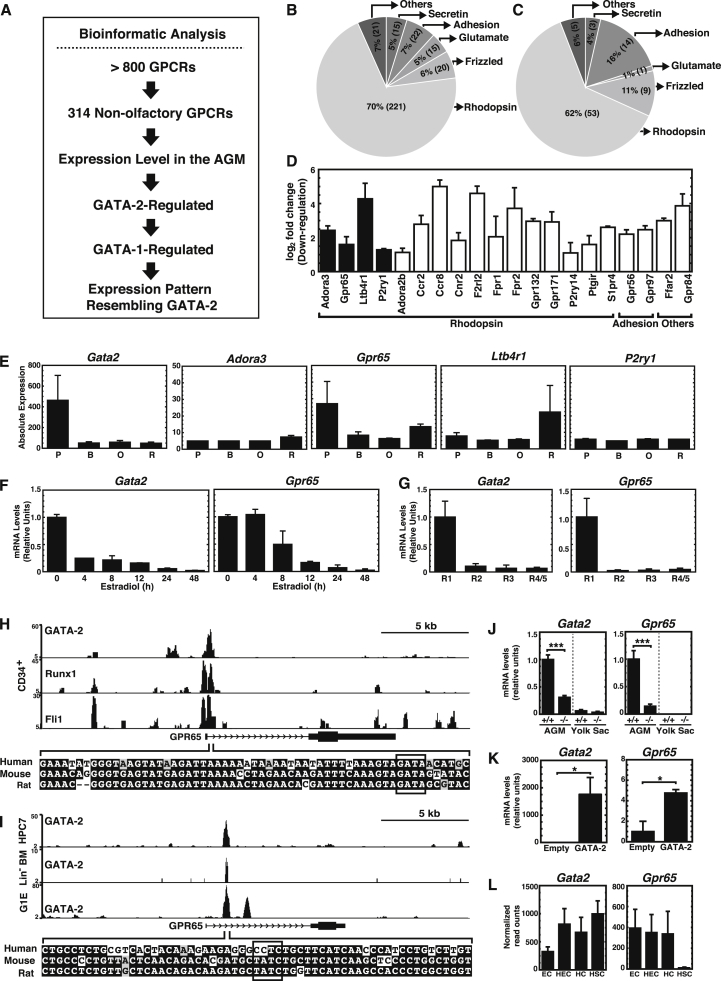
Figure 3.
Global GPCR Analysis in the AGM
(A) Global GPCR analysis.
(B) Non-olfactory GPCRs (N = 314) were categorized into Secretin, Adhesion, Glutamate, Frizzled/Taste2, and Rhodopsin families based on sequence homology.
(C) Classification of 85 GPCRs expressed in the AGM (>5 transcripts per million) into five families.
(D) Bar graph depicting GATA-2-regulated genes from RNA-seq analysis of +9.5+/+ and +9.5−/− AGMs (Gao et al., 2013). Black bars, genes co-regulated by GATA-1 according to our prior microarray analysis of G1E-ER-GATA with or without β-estradiol treatment (DeVilbiss et al., 2013).
(E) Gata2, Adora3, Gpr65, Ltb4r1, and P2ry1 expression during erythropoiesis. B, basophilic erythroblast; O, polyorthochromatic erythroblast; P, proerythroblast; R, reticulocyte (http://www.cbil.upenn.edu/ErythronDB/).
(F) Time course of Gata2 and Gpr65 expression following β-estradiol treatment in G1E-ER-GATA cells (n = 3 independent experiments).
(G) RT-PCR analysis of Gata2 and Gpr65 in FACS-sorted R1, R2, R3, and R4/5 populations from fetal liver (n = 3 independent experiments).
(H and I) ChIP signal map for Gpr65 in human CD34 cells (H) (Beck et al., 2013), mouse HPC7 cells (Wilson et al., 2010), Lin− bone marrow cells (Li et al., 2011), and G1E cells (Trompouki et al., 2011) (I).
(J and K) RT-PCR analysis of Gata2 and Gpr65 mRNA in +9.5+/+ and +9.5−/− AGM (5 litters: +9.5+/+ [n = 8 embryos]; +9.5−/− [n = 6 embryos]) and yolk sac (three litters: +9.5+/+ [n = 7 embryos]; +9.5−/− [n = 5 embryos]) (J), and MAE cells expressing GATA-2 (K) (n = 3 independent experiments).
(L) RNA-seq of Gata2 and Gpr65 mRNA in FACS-sorted endothelial cells (EC), hemogenic endothelial cells (HEC), hematopoietic cells (HC), and hematopoietic stem cells (HSC) from the AGM (Solaimani Kartalaei et al., 2015).
Error bars represent SEM. ∗p < 0.05; ∗∗∗p < 0.001 (two-tailed unpaired Student's t test).