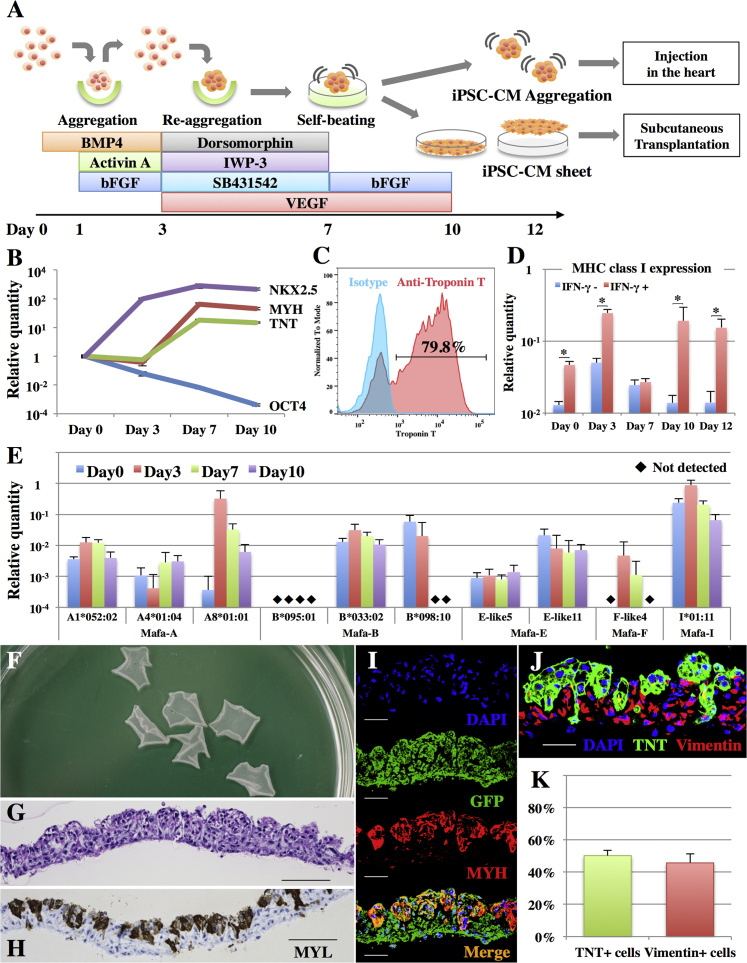
Figure 2.
Generation of Cynomolgus Macaque iPSC-CM Sheets
(A) Cardiomyogenic differentiation protocol and the generation of iPSC-CM sheets.
(B) Expression of NKX2.5, MYH, TNT, and OCT4 transcripts in iPSCs on days 0, 3, 7, and 10 as analyzed by real-time PCR. Results are relative to those at day 0 and are expressed as the means ±SD (n = 3 independent experiments).
(C) Representative flow cytometry data of iPSC-CMs at day 10, stained with anti-troponin T antibodies or the isotype control.
(D) Expression of MHC class I genes in iPSCs on days 0, 3, 7, 10, and 12 with or without IFN-γ stimulation as analyzed by real-time PCR. Results are relative to those of the peripheral blood and are expressed as the means ±SD (n = 3 independent experiments). ∗p < 0.05.
(E) Expression of MHC class I genes in iPSCs on days 0, 3, 7, and 10 as analyzed by real-time PCR. Mafa-A, B, F, E, and I are the MHC-A, B, F, E, and I genes, respectively, in cynomolgus macaques. The relative quantities of each allele are compared with those of the peripheral blood and are expressed as the means ±SD (n = 3 independent experiments).
(F) Macaque iPSC-CM sheets in a 10-cm dish.
(G) H&E staining of the iPSC-CM sheet. Scale bar, 100 μm.
(H) Immunohistochemistry of MYL in the iPSC-CM sheet. Scale bar, 100 μm.
(I) Immunohistochemistry of GFP (Alexa Fluor 488), MYH (Alexa Fluor 546), and DAPI in the iPSC-CM sheet. Scale bar, 30 μm.
(J) Immunohistochemistry of TNT (Alexa Fluor 488), vimentin (Alexa Fluor 546), and DAPI in the iPSC-CM sheet. Scale bar, 30 μm.
(K) Percentage of TNT- and vimentin-positive cells in the iPSC-CM sheet as analyzed by flow cytometry (representative data are shown in Figure S1C). Results are expressed as the means ±SD (n = 5 independent experiments).