Fig. 3.
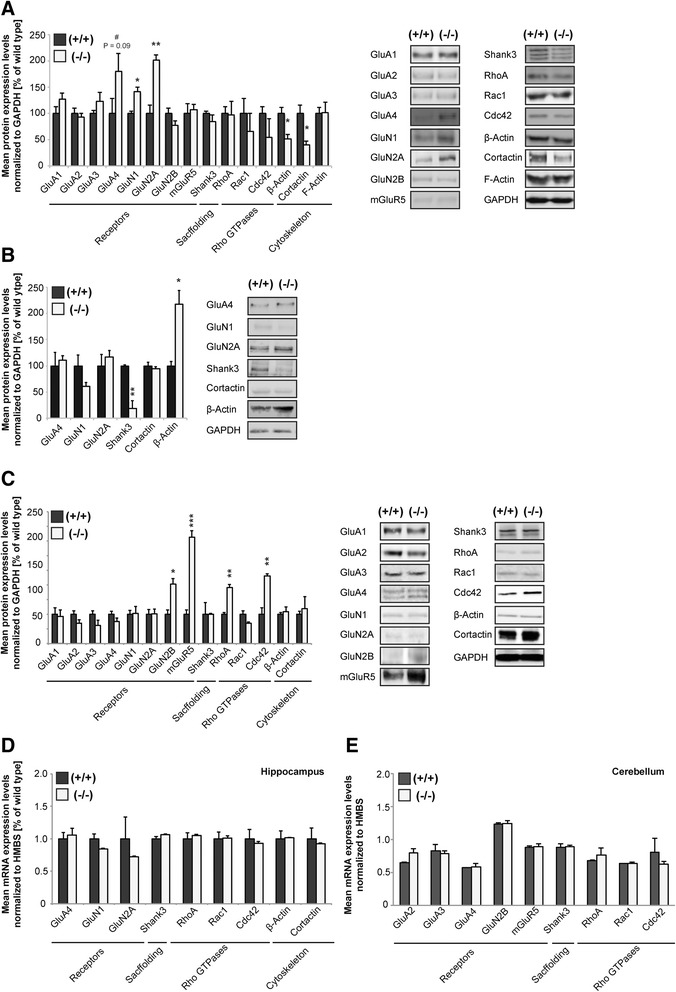
Altered synapse composition and function in RICH2−/− mice. a Western blot analysis (showing relative percentages of mean + SEM) of hippocampal synapse-enriched P2-fractions extracted from P70 wild type (+/+, n = 3), and knock-out (−/−, n = 3) mice. Proteins were normalized to GAPDH expression levels. Right panel: Representative illustration from hippocampal P2-immunoblots. For each protein analyzed two representative immunonblot-signals are illustrated per genotype. A significant increase can be seen for GluN1 and GluN2A levels, while a significant decrease is visible for β-Actin and Cortactin (unpaired t-test, GluN1: df = 4, p = 0.0102, t = 4.4439; GluN2A: df = 4, p = 0.005, t = 5.5458; β – Actin: df = 4, p = 0.0259, t = 3.463; Cortactin: df = 4, p = 0.0264, t = 3.3999). b Western blot analysis (showing relative percentages of mean + SEM) of hippocampal S2-fractions extracted from P70 wild type (+/+, n = 3), and knock-out (−/−, n = 3) mice. Proteins were normalized to GAPDH expression levels. Right panel: Representative illustration from hippocampal S2-immunoblots. For each protein analyzed two representative immunonblot-signals are illustrated per genotype. c Western blot analysis (showing relative percentages of mean + SEM) of cerebellar synapse-enriched P2-fractions extracted from P70 wild type (+/+, n = 3), and knock-out (−/−, n = 3) mice. Proteins were normalized to GAPDH expression levels. Right panel: Representative illustration from cerebellar P2-immunoblots. For each protein analyzed two representative immunonblot-signals are illustrated per genotype. A significant increase in protein expression levels can be seen for GluN2B, mGluR5, RHOA, and CDC42 (unpaired t-test, GluN2B: df = 4, p = 0.0126, t = 4.2997; mGluR5: df = 4, p = 0.0006, t = 10.0161; RhoA: df = 4, p = 0.0011, t = 8.4287; CDC42: df = 4, p = 0.005, t = 5.5827). Note that expressions of GluN2B and mGluR5 in general were found to be very low in cerebellar lysates. d, e qRT-PCR analysis showing relative changes in hippocampal and cerebellar mRNA-levels (normalized to HMBS) using total RNA extracted from crude cellular homogenate from P70 wild type (+/+, n = 3) and knock-out (−/−, n = 3) mice. Each qRT-PCR experiment was set up of 3 biological and 3 technical replicates. The results show no significant differences of tested genes between wild type and RICH2−/− mice in hippocampus (unpaired t-test, GluA4: df = 4, p = 0.7060, t = 0.105956; GluN1: df = 4, p = 0.1175, t = 1.9897; GluN2A: df = 4, p = 0.4582, t = 0.8201; Shank3: df = 4, p = 0.1658, t = 1.6924; Cortactin: df = 4, p = 0.6827, t = 0.4399; β actin: df = 4, p = 0.9026, t = 0.1303; RhoA: df = 4, p = 0.593, t = 0.58; CDC42: df = 4, p = 0.6591, t = 0.4756; Rac1: df = 4, p = 0.931, t = 0.0922) (d) and cerebellum (GluA2: df = 4, p = 0.0836, t = 2.2929; GluA3: df = 4, p = 0.7140, t = 0.3936; GluA4: df = 4, p = 0.7553, t = 0.3338; GluN2B: df = 4, p = 0.8972, t = 0.1377; mGluR5: df = 4, p = 0.8873, t = 0.1509; Shank3: df = 4, p = 0.9134, t = 0.1158; RhoA: df = 4, p = 0.4049, t = 0.9303; CDC42: df = 4, p = 0.4359, t = 0.8649; Rac1: df = 4, p = 0.9799, t = 0.0268) (e)
