Fig. 1.
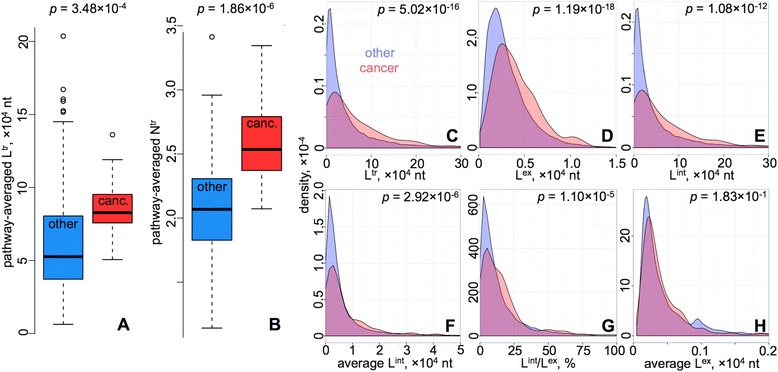
Enrichment of long transcripts and genes with greater number of transcript variants in cancer pathways. a Distribution of the pathway-averaged Ltr transcript length in cancer (red) and other (blue) pathways. b Distribution of the pathway-averaged Ntr number of transcripts in cancer (red) and other (blue) pathways. In the boxplots (a, b), each box is constructed via the median, first and third quartiles of the distribution. The whiskers show the range of values that are within the 1.5 times IQR (interquartile range). Individual points indicate the outliers. c-h Distributions of gene length and exon/intron composition descriptors in cancer (red) and other (blue) pathways. The plots are for the Ltr transcript (exons, UTR inclusive, and introns) length (c), Lex summed exon length (d), Lint summed intron length (e), average Lint length of a single intron (f), Lint/Lex summed intron to summed exon length ratio (g) and average Lex length of a single exon (h) for all the genes in other and cancer pathways. The p-values quantifying the significance of a positive shift in the distributions for the cancer pathways, as compared to others, are shown on top of each plot. Data from each of the c-h plots come from 380 cancer-linked and 18839 other genes
