Fig. 9.
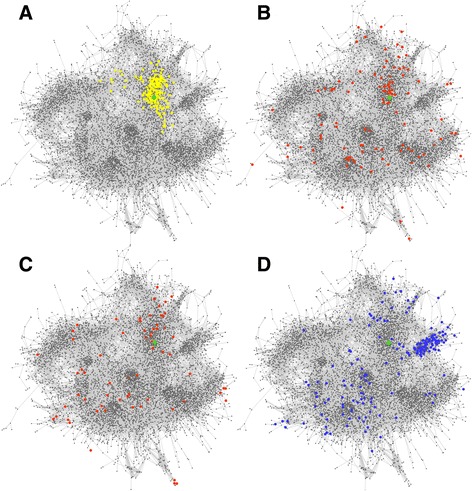
Experimental Cross-validation of the Predictive Power of ChlamyNET using RNA-seq Data from Algae Overexpressing the CrDOF gene. a The CrDOF gene (identified as a green diamond in ChlamyNET) has a neighbourhood at distance two consisting of 216 genes represented in yellow. These genes showed an average fold-change increase of 2.7 which is significantly higher than the fold-change in the rest of ChlamyNET with a p-value of 5.63° 10 −3. b Genes increasing their expression level in LD conditions at least by a four fold-change in the CrDOF genotype when compared to the wild type CW15 are represented in red. Note that the neighbourhood of the CrDOF gene, represented in green, is enriched in these genes according to a p-value of 0.029 obtained using Fisher's exact test. c Genes increasing their expression level in SD conditions at least by a four fold-change in the CrDOF genotype when compared to the wild type CW15 are represented in red. These genes tend to group around the CrDOF gene, represented in green. d Inhibited genes in LD conditions in the CrDOFin genotype when compared to the wild type CW15 with at least by a four fold-change are represented in blue. Note that cluster 2 (brown) involved in DNA replication and cell cycle processes is significantly enriched in these genes
