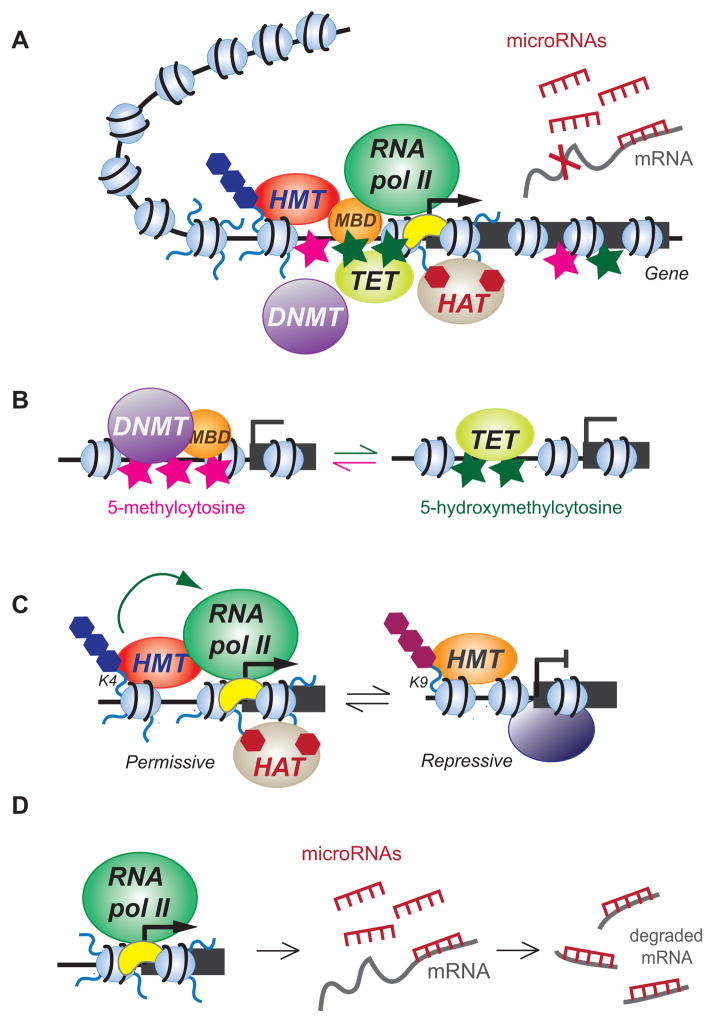
Figure 2.
Several epigenetic mechanisms relevant to the effects of exogenous cannabinoids. (A) Gene expression is regulated by a network of DNA elements (e.g. promoters) and trans-acting factors (proteins that bind to the DNA) that interact physically and functionally to generate appropriate mRNA transcript levels from a gene. The resulting balance can be disrupted by drug exposure. Regulatory mechanisms include DNA methylation (Me), positioning and post-translational modifications of nucleosomes (small blue balls), recruitment of sequence-specific and basal transcription factors and RNA polymerase II, and non-coding RNAs. The DNA-protein structure forms three-dimensional structures (represented by the chromatin loop) that influence the expression of associated genes. (B) DNA methyltranserases (DNMT) generate 5-methylcytosine (pink stars) at CpG sites, facilitated by methyl-CpG binding domain (MBD)-containing proteins. Ten-eleven translocation (TET) proteins mediate the oxidation of 5-methylcytosine to 5-hydroxymethylcytosine (green stars), leading to demethylation of the DNA. (C) Modifications of nucleosomal histone tails such as methylation (Me) and acetylation (Ac) are mediated by histone methyltransferases (HMT) and histone acetyltransferases (HAT), respectively. Depending on modified amino acid residue, methylation can have either permissive (e.g. on lysine4, K4) or repressive (e.g. on lysine 9, K9) effects on transcription. Permissive modifications facilitate gene activation via the recruitment of the RNA polymerase II machinery. Acetylation is removed by histone deacetylases (HDAC) and can lead to transcriptional repression. (D) MicroRNAs are produced from specific genes and target protein-coding messenger RNAs (mRNA) for degradation, thereby prevention protein production.