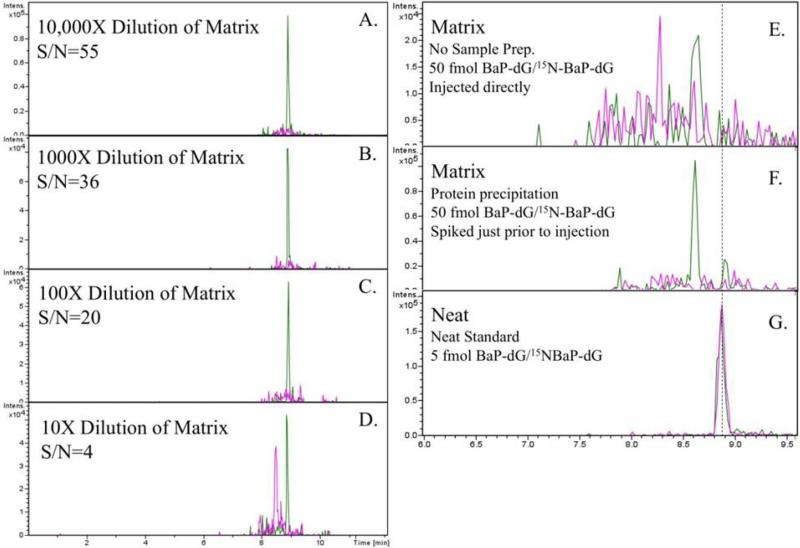
Figure 8.
DNA digests were enriched on the HPLC chip by online SPE. Here the washing time (and volume) is varied to determine the effect on analyte signal. Flow rate is 4 μL/min with 30% methanol in water containing 0.1% acetic acid. Data show that the analyte begins to elute (to waste) after 6 minutes (24 μL) or more of enrichment. Note the enrichment column has a volume of 40 nL, therefore 6 minutes of washing equals 600 column volumes. Also note that the separation gradient was started after enrichment, therefore the increasing retention time observed is artificial (BaP-dG elutes at 45% B in each case).