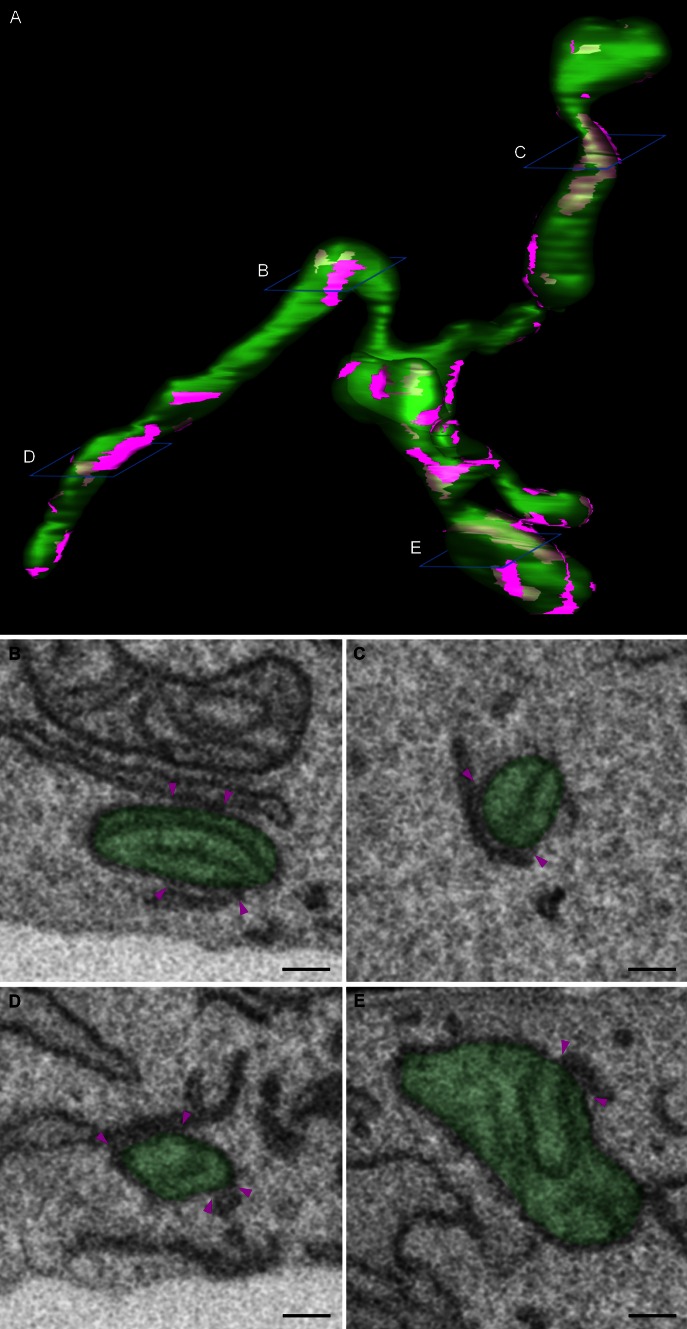
Fig. 1.
3D morphology of ER-mitochondria contact sites. Mouse embryonic fibroblasts were imaged using a Zeiss Auriga Crossbeam focused ion beam scanning electron microscope (FIB-SEM). By automated serial block face imaging, large image stacks are generated at high resolution, allowing precise 3D reconstruction of a large cellular volume. In this dataset, over 600 10 nm z-slices were obtained resulting in a 3D dataset at 5 × 5 × 10 nm3 voxels. Mitochondria and their contact sites with the endoplasmic reticulum (ER) were manually segmented and 3D reconstructed in IMOD (http://bio3d.colorado.edu/imod/). A video of this dataset and the reconstruction can be seen in Supplementary movie 1. a The complete reconstruction of two mitochondria (transparent green) and their ER-mitochondria contact sites (magenta) is shown. A contact site is defined as a region where the ER and the mitochondrial membranes are in closer proximity than 30 nm. It is clear that a single mitochondrion makes multiple contacts with the ER and that these contacts are diverse in size, ranging from punctate sites to large patches of the outer mitochondrial membrane being juxtaposed to the ER. b–e Represent different examples of scanning electron micrographs extracted from the volume illustrating a section of the mitochondria and their contacts with the ER. The reconstructed mitochondria depicted in a are shown in transparent green. Magenta arrowheads mark the borders of the ER-mitochondria contact sites. The position of these slices is depicted in blue in a. Scale bar 200 nm