Abstract
Background
Up to now, fecal–oral and oral–oral are the most commonly known routes for transmission of H. pylori, therefore, contaminated water can play an important role in transmission of H. pylori to humans. Genotyping using virulence markers of H. pylori is one of the best approaches to study the correlations between H. pylori isolates from different samples. The present research was carried out to study the vacA, cagA, cagE, oipA, iceA and babA2 genotyping and antimicrobial resistance properties of H. pylori isolated from the bottled mineral water samples of Iran.
Results
Of 450 samples studied, 8 samples (1.77 %) were contaminated with H. pylori. Brand C of bottled mineral water had the highest prevalence of H. pylori (3.63 %). The bottled mineral water samples of July month had the highest levels of H. pylori-contamination (50 %). H. pylori strains had the highest levels of resistance against metronidazole (62.5 %), erythromycin (62.5 %), clarithromycin (62.5 %), amoxicillin (62.5 %) and trimethoprim (62.5 %). Totally, 12.5 % of strains were resistant to more than 6 antibiotics. VvacAs1a (100 %), vacAm1a (87.5 %), cagA (62.5 %), iceA1 (62.5 %), oipA (25 %), babA2 (25 %) and cagE (37.5 %) were the most commonly detected genotypes. M1as1a (62.5 %), m1as2 (37.5 %), m2s2 (37.5 %) and S1a/cagA+/IceA2/oipA-/babA2-/cagE- (50 %) were the most commonly detected combined genotypes.
Conclusions
Contaminated bottled mineral water maybe the sources of virulent and resistant strains H. pylori. Careful monitoring of bottled mineral water production may reduce the risk of H. pylori transmission into the human population.
Keywords: Helicobacter pylori, Genotyping, Antibiotic resistance properties, Bottled mineral water, Iran
Background
In a day, Millions of people use drinking water and in some areas in which the quality of tap drinking water is low, consumption of bottled mineral water is common. Bottled mineral waters are a rich sources of trace elements copper, zinc, and iron and also magnesium, fluorine, sodium, and calcium. Their consumption is routine in hotels, restaurants, airplanes and other vehicles and also medical practitioners usually prescribe these kinds of waters for patients with malnutrition. Low risk of water-borne pathogens and chemical toxins exist for bottled drinking water, increasing their consumption over the time. In keeping with this, there are some reports regarding the contamination of these kinds of water with dangerous pathogens such as Helicobacter pylori, Vibrio cholera, Salmonella typhimurium and Escherichia coli [1, 2].
H. pylori is a Gram negative, micro-aerophilic and flagellated bacterium which is known as a causative agent of gastric adenocarcinoma, type B gastritis and mucosa-associated lymphoid tissue lymphoma [3, 4]. In fact, H. pylori colonization in gastric mucosa is the primary cause of ulcers in the stomach and duodenum [4]. Documented data revealed that near 50 % of world populations have been infected with H. pylori [3, 5, 6]. In keeping with this, its exact routes of transmission and origin are still unknown [7]. Based on the fecal–oral and oral–oral transmission routes of the H. pylori, contaminated water can play an important role in spread of H. pylori to humans [8, 9].
Colonization and invasion of H. pylori to gastric mucous is dependent on a number of virulence factors. Some of the most important virulence factors of this bacterium are the vacuolating cytotoxin (vacA), induced by contact with the epithelium antigen (iceA), cytotoxin associated gene (cag), blood group antigen-binding adhesion (babA) and outer inflammatory protein (oip) [10–13]. The vacA gene is polymorphic, comprising variable signal regions (type s1 or s2) and mid-regions (type m1 or m2). The s1 type is further subtyped into s1a, s1b and s1c subtypes and the m1 into m1a and m1b subtypes. The iceA gene has two main allelic variants iceA1 and iceA2 but their functions are not yet clear. The cagA gene has been detected in the specimens taken from the severe cases of peptic ulcer [10–13]. CagE is 1 of 6 genes located within the pathogenicity island shown to induce secretion of chemokines, such as interleukin IL-8, and induce inflammation [14]. The oipA gene plays a significant role in effective colonization of bacteria into the mucosa [15]. Genotyping using these virulence markers is considered as one of the best approaches for the study of relationships between H. pylori isolates from different samples.
Efficient antibiotic therapy is one of the best approaches for treatment of infectious diseases transmitted through water. Water-borne H. pylori infections are no exception to this principle. However, therapeutic protocols have become somewhat limited because of the presence of multidrug resistant strains of this bacterium [12, 13].
Consumption of bottled mineral water is so common among Iranian people. Besides, the prevalence of H. pylori is considerable among Iranian sources of infection and other sites of the wold [11, 16–20]. Therefore, study the important role of contaminated water as a risk factor for transmission of H. pylori is essential. These important points and also lack of community studies in relation to antibiotic resistance pattern of water-borne H. pylori,caused us to carried out the present study in order to investigate the exact status of vacA, cagA, iceA, oipA, cage and baba2 genotypes status and antibiotic resistance pattern of H. pylori isolates of Iranian bottled mineral water samples.
Results
A total of 450 bottled mineral water samples were examined for the presence of H. pylori, its genotypes, and its antimicrobial resistance properties. Table 1 shows the total distribution of H. pylori in the bottled mineral water samples of 4 different brands and various seasons from Iran. Of 450 bottled mineral water samples collected, 8 samples (1.77 %) were contaminated with H. pylori. The results of culture methods were confirmed using the 16S rRNA-based PCR. The water samples of brand C had the highest prevalence (3.63 %) of H. pylori, while those of brand A had the lowest (0.83 %). A statistically significant difference was seen between the distributions of H. pylori in different brands of bottled mineral water (P < 0.05). We found that the samples which were collected in the summer season had the highest prevalence of bacteria (4.54 %). Significant statistical difference was seen for the prevalence of H. pylori between warm and cold seasons of the year (P < 0.05). Monthly distribution of H. pylori in the bottled mineral water samples is shown in Fig. 1. We found that the bottled mineral water samples from July had the highest levels of H. pylori-contamination (50 %).
Table 1.
Total distribution of Helicobacter pylori in the tested bottled mineral water samples
| Type of bottled mineral water | No. samples collected | No. Helicobacter pylori isolates (%) | No. isolates confirmed using 16SrRNA based-PCR (%) | |
|---|---|---|---|---|
| Brands | A | 120 | 1 (0.83)a | 1 (0.83)a |
| B | 110 | 1 (0.90)a | 1 (0.90)a | |
| C | 110 | 4 (3.63)b | 4 (3.63)b | |
| D | 110 | 2 (1.81)a | 2 (1.81)a | |
| Total | 450 | 8 (1.77) | 8 (1.77) | |
| Seasons | Spring | 112 | 1 (0.89)a | 1 (0.89)a |
| Summer | 110 | 6 (4.54)b | 6 (4.54)b | |
| Autumn | 113 | 1 (0.88)a | 1 (0.88)a | |
| Winter | 115 | - | - | |
| Total | 450 | 8 (1.77) | 8 (1.77) | |
a, bDissimilar letters in each column shows significant differences about P <0.05
Fig. 1.
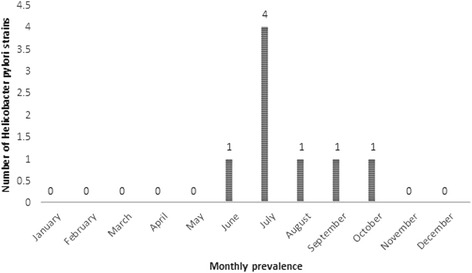
Monthly prevalence of Helicobacter pylori in tested bottled mineral water samples
The results of antimicrobial resistance pattern of H. pylori isolates of tested bottled mineral water samples is shown in Table 2. H. pylori strains of bottled mineral water harbored the highest levels of resistance against ampicillin (75 %), followed by metronidazole (62.5 %), erythromycin (62.5 %), clarithromycin (62.5 %), amoxicillin (62.5 %) and trimethoprim (62.5 %) antimicrobial agents. Prevalence of antibiotic resistance against moxifloxacin, tinidazole and ciprofloxacin were 25, 50 and 50 %, respectively. There were statistically significant differences in the levels of antibiotic resistance between ampicillin and cefsulodin (P =0.013), ampicillin and furazolidone (P =0.018), clarithromycin and furazolidone (P =0.029) and ampicillin and moxifloxacin (P =0.027). Prevalence of multidrug resistant of H. pylori strains of bottled mineral water samples is shown in Fig. 2. All of the 8 strains of H. pylori were resistant to at least one of the tested antibiotics. The highest levels of resistance was found only for one isolate which was resistant to 10 antibiotics.
Table 2.
Antimicrobial resistance pattern of Helicobacter pylori isolates of tested bottled mineral water samples
| Brands (No. positive) | Pattern of antibiotic resistance (%) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AM10* | Met5 | ER5 | CLR2 | AMX 10 | Lev5 | Cef30 | TRP25 | FZL1 | Mox5 | Tin4 | CIP5 | |
| A (1) | 1 (100)a | 1 (100)a | 1 (100)a | 1 (100)a | 1 (100)a | - | - | - | - | - | - | - |
| B (1) | 1 (100)a | 1 (100)a | 1 (100)a | 1 (100)a | 1 (100)a | 1 (100)a | - | 1 (100)a | - | - | 1 (100)a | 1 (100)a |
| C (4) | 2 (50)a | 2 (50)a | 2 (50)a | 2 (50)a | 2 (50)a | 2 (50)a | 1 (25)b | 3 (75)a | 1 (25)b | 1 (25)b | 2 (50)a | 2 (50)a |
| D (2) | 2 (100)a | 1 (50)a | 1 (50)a | 1 (50)a | 1 (50)a | 1 (50)a | 1 (50)b | 1 (50)a | 1 (50)b | 1 (50)b | 1 (50)a | 1 (50)a |
| Total (8) | 6 (75)a, * | 5 (62.5)a | 5 (62.5)a | 5 (62.5)a | 5 (62.5)a | 4 (50)a | 2 (25)b | 5 (62.5)a | 2 (25)b | 2 (25)b | 4 (50)a | 4 (50)a |
*AM10: ampicillin (10 μg), Met5: metronidazole (5 μg), ER5: erythromycin (5 μg), CLR2: clarithromycin (2 μg), AMX10: amoxicillin (10 μg), Lev5: levofloxacin (5 μg), Cef30: cefsulodin (30 μg), TRP25: trimethoprim (25 μg), FZL1: furazolidone (1 μg), Mox5: moxifloxacin (5 μg), Tin: tinidazole (prepared from pure powders, Sigma, 5 μg) and CIP5: ciprofloxacin (5 μg)
a, bDissimilar letters in each row shows significant differences about P <0.05
Fig. 2.

Prevalence of multidrug resistant strains of Helicobacter pylori isolated from tested bottled mineral water samples
Distribution of various genotypes of vacA alleles, cagA, iceA1, iceA2, oipA, cagE and babA2 in the H. pylori isolates of bottled mineral water is shown in Table 3. The most commonly detected genotypes amongst the H. pylori isolates of bottled mineral water samples were vacAs1a (100 %), vacAm1a (87.5 %), cagA (62.5 %) and iceA1 (62.5 %). The prevalence of oipA, babA2 and cagE genotypes were 25, 25 and 37.5 %, respectively. The distribution of combined genotypes of H. pylori isolates is shown in Table 4. The most commonly detected combined genotypes were m1as1a (62.5 %), m1as2 (37.5 %) and m2s2 (37.5 %). Significant differences were found between the incidence of genotypes and brands of bottled mineral water samples (P <0.05). Forty seven different combined genotypes were detected in the H. pylori strains of bottled mineral water (Table 5). The most commonly detected combined genotypes were S1a/cagA+/IceA2/oipA-/babA2-/cagE- (50 %), M2/cagA+/IceA1/oipA-/babA2-/cagE- (37.5 %), S1a/cagA-/IceA1/oipA-/babA2-/cagE- (37.5 %), S2/cagA+/IceA1/oipA-/babA2-/cagE- (37.5 %) and M1a /cagA+/IceA2/oipA-/babA2-/cagE- (37.5 %).
Table 3.
Total distribution of various genotypes in Helicobacter pylori strains of tested bottled mineral water samples
| Brands (No. positive) | Distribution of various genotypes (%) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S1a | S1b | S1c | S2 | M1a | M1b | M2 | CagA | IceA1 | IceA2 | OipA | BabA2 | cagE | |
| A (1) | 1 (100) | - | - | 1 (100) | 1 (100) | - | 1 (100) | 1 (100) | 1 (100) | - | - | - | 1 (100) |
| B (1) | 1 (100) | - | - | - | 1 (100) | - | 1 (100) | 1 (100) | 1 (100) | - | - | 1 (100) | 1 (100) |
| C (4) | 4 (100) | 1 (25) | - | 2 (50) | 4 (100) | 1 (25) | 1 (25) | 2 (50) | 2 (50) | 1 (25) | 1 (25) | 1 (25) | 1 (25) |
| D (2) | 2 (100) | 1 (50) | - | 1 (50) | 1 (50) | 1 (50) | 1 (50) | 1 (50) | 1 (50) | - | 1 (50) | - | - |
| Total (8) | 8 (100) | 2 (25) | - | 4 (50) | 7 (87.5) | 2 (25) | 4 (50) | 5 (62.5) | 5 (62.5) | 1 (12.5) | 2 (25) | 2 (25) | 3 (37.5) |
Table 4.
Distribution of combined genotypes of Helicobacter pylori isolated from tested bottled mineral water samples
| Genotypes | Prevalence (%)* |
|---|---|
| M1as1a | 5 (62.5) |
| M1as1b | 1 (12.5) |
| M1bs1a | 1 (12.5) |
| M1bs1b | - |
| M1as1c | - |
| M1bs1c | - |
| M2s1a | 2 (25) |
| M2s1b | 1 (12.5) |
| M2s1c | - |
| M2s2 | 3 (37.5) |
| M1as2 | 3 (37.5) |
| M1bs2 | 1 (12.5) |
| M1am2 | 2 (25) |
| M1bm2 | - |
| CagA+ | 5 (62.5) |
| CagA- | 3 (37.5) |
| CagE+ | 3 (37.5) |
| CagE- | 5 (62.5) |
| IceA1 | 5 (62.5) |
| IceA2 | 1 (12.5) |
| IceA1 IceA2 | - |
| OipA+ | 2 (25) |
| OipA- | 6 (75) |
| babA2+ | 2 (25) |
| babA2- | 6 (75) |
*From a total of 8 positive strains of H. pylori
Table 5.
Combined vacA, cagA, cagE, iceA, babA2 and oipA genotypes of Helicobacter pylori isolated from tested mineral water samples
| Combined genotype* | Frequency (%) |
|---|---|
| S1a/cagA+/IceA1/oipA+/babA2+/cagE+ | 1 (12.5) |
| S1a/cagA+/IceA1/oipA-/babA2+/cagE+ | 2 (25) |
| S1a/cagA-/IceA1/oipA-/babA2+/cagE+ | 1 (12.5) |
| S1a/cagA+/IceA1/oipA+/babA2-/cagE+ | 2 (25) |
| S1a/cagA+/IceA1/oipA+/babA2+/cagE- | 1 (12.5) |
| S1a/cagA+/IceA1/oipA+/babA2-/cagE- | 2 (25) |
| S1a/cagA-/IceA1/oipA-/babA2-/cagE- | 3 (37.5) |
| S1a/cagA+/IceA1/oipA+/babA2-/cagE- | 2 (25) |
| S1a/cagA+/IceA1/oipA-/babA2 + -/cagE- | 2 (25) |
| S1a/cagA+/IceA2/oipA-/babA2-/cagE- | 4 (50) |
| S1b/ cagA+/IceA1/oipA+/babA2-/cagE+ | 1 (12.5) |
| S1b/ cagA+/IceA1/oipA+/babA2+/cagE- | 1 (12.5) |
| S1b/ cagA+/IceA1/oipA-/babA2-/cagE- | 2 (25) |
| S2/cagA-/IceA1/oipA+/babA2+/cagE+ | 1 (12.5) |
| S2/cagA+/IceA1/oipA-/babA2+/cagE+ | 1 (12.5) |
| S2/cagA-/IceA1/oipA-/babA2+/cagE+ | 1 (12.5) |
| S2/cagA+/IceA1/oipA+/babA2-/cagE+ | 1 (12.5) |
| S2/cagA+/IceA1/oipA+/babA2+/cagE- | 1 (12.5) |
| S2/cagA+/IceA1/oipA+/babA2-/cagE- | 1 (12.5) |
| S2/cagA+/IceA1/oipA-/babA2+/cagE- | 2 (25) |
| S2/cagA+/IceA1/oipA-/babA2-/cagE+ | 2 (25) |
| S2/cagA+/IceA1/oipA-/babA2-/cagE- | 3 (37.5) |
| M1a/ cagA+/IceA1/oipA+/babA2+/cagE+ | 1 (12.5) |
| M1a/cagA-/IceA1/oipA+/babA2+/cagE+ | 1 (12.5) |
| M1a /cagA+/IceA1/oipA-/babA2+/cagE+ | 1 (12.5) |
| M1a /cagA-/IceA1/oipA-/babA2+/cagE+ | 1 (12.5) |
| M1a /cagA+/IceA1/oipA+/babA2-/cagE+ | 1 (12.5) |
| M1a /cagA+/IceA1/oipA+/babA2+/cagE- | 2 (25) |
| M1a /cagA+/IceA1/oipA+/babA2-/cagE- | 2 (25) |
| M1a /cagA-/IceA1/oipA-/babA2-/cagE- | 2 (25) |
| M1a /cagA+/IceA1/oipA+/babA2+/cagE- | 1 (12.5) |
| M1a /cagA+/IceA1/oipA-/babA2-/cagE+ | 2 (25) |
| M1a /cagA+/IceA1/oipA-/babA2+/cagE- | 2 (25) |
| M1a /cagA+/IceA2/oipA-/babA2-/cagE- | 3 (37.5) |
| M1b/ cagA+/IceA1/oipA-/babA2+/cagE+ | 1 (12.5) |
| M1b / cagA+/IceA1/oipA+/babA2-/cagE+ | 1 (12.5) |
| M1b / cagA+/IceA1/oipA+/babA2+/cagE- | 1 (12.5) |
| M1b / cagA+/IceA1/oipA-/babA2-/cagE- | 1 (12.5) |
| M2/cagA+/iceA1/oipA+/babA2+/cagE+ | 1 (12.5) |
| M2/cagA-/IceA1/oipA+/babA2+/cagE+ | 1 (12.5) |
| M2/cagA+/IceA1/oipA-/babA2+/cagE+ | 1 (12.5) |
| M2/cagA-/IceA1/oipA-/babA2+/cagE+ | 1 (12.5) |
| M2/cagA+/IceA1/oipA+/babA2-/cagE+ | 1 (12.5) |
| M2/cagA+/IceA1/oipA+/babA2+/cagE- | 1 (12.5) |
| M2/cagA+/IceA1/oipA+/babA2-/cagE- | 2 (25) |
| M2/cagA+/IceA1/oipA-/babA2+/cagE- | 2 (25) |
| M2/cagA+/IceA1/oipA-/babA2-/cagE- | 3 (37.5) |
*From a total of 8 positive strains of H. pylori
Discussion
The present research was carried out to study the prevalence of H. pylori in the bottled mineral water samples of 4 major brands in Iran as well as to determine the distribution of vacA, cagA, cagE, iceA, oipA and babA2 genotype status and antibiotic resistance patterns of H. pylori isolates. To our best knowledge, this is the first prevalence report of genotyping and antimicrobial resistance properties of H. pylori in the bottled mineral water samples in the world. We found that 1.77 % of bottled mineral water samples were contaminated with resistant and virulent strains of H. pylori. With respect to the high accuracy which were performed on the producing of mineral water, our results reported a significant public health concern.
In contrast to findings of our investigation, Watson et al. (2004) [21] failed to isolate H. pylori from drinking water samples of United Kingdom. Isolation of H. pylori using the culture media is important due to the necessity of detecting viable bacteria in water. Previous investigation in Iran [1] showed that the distribution of H. pylori in various types of water samples including tap water, dental units’ water and water cooler of public places had the ranges of 2–6 %, which was higher than our results. Nurgalieva et al. (2002) [22] revealed that river water was a high risk factor for H. pylori infection in Kazakhstan. Accordingly, they stated that transmission of H. pylori could be waterborne. Drinking water can pose a substantial threat for transmission of H. pylori because of several important criteria [23]. These criteria include the ability of H. pylori to adhere to different materials and to co-aggregate with other bacteria and form complex structures on pipes or other surfaces in contact with water [23]. Notion about the disability of H. pylori to survive alone in running water, but to develop a symbiotic relationship and form complex structures on contact surfaces [24], makes it rational to assume that groundwater is a reservoir for H. pylori due to its stagnant nature. According to the high application of groundwater in the production of bottled mineral water, it is not surprising that 1.77 % of our samples were positive for this bacterium. Several investigations have shown a considerable prevalence of H. pylori in the ground water samples between years 1999–2007 in United Sates [25–27] and Japan [28].
Other possible reasons for the high prevalence of H. pylori in bottled mineral water samples in Iran are a) lack of competent approaches for water sanitization b) expending the distrustful groundwater for producing the bottled mineral water c) the opportunity of presence of bacterial colonies as a biofilm in the in pipes used for water transfer d) the opportunity of leakage of household, industrial and agricultural wastewater to the sources of mineral water and finally e) lack of personal hygiene of refinery room’s staffs. According to the high prevalence of H. pylori in bottled mineral water samples, it can be concluded that the food safety regulations as well as quality standards—including good manufacturing practices (GMPs), good agricultural practices (GAPs) and hazard analysis and critical control points (HACCP)—are not introduced for Iranian mineral water factories.
Marked monthly distribution with the higher prevalence in July month was seen in the H. pylori strains of bottled mineral water. The main reason for the higher prevalence of H. pylori in summer is the fact that during these times of the year climatic events including temperature, rain and barometric pressure may have influence on the prevalence of this bacterium. Bacteria have the higher growth and occurrence in warm condition. Yahaghi et al. (2014) [13] reported a similar seasonal distribution for the H. pylori strains of vegetable and salad samples. They showed statistically significant differences for the incidence of H. pylori between hot and cold seasons of the year.
Consumption of contaminated bottled mineral water of our study may lead to the gastrointestinal disorders, because of the presence of resistant and virulent strains of H. pylori. All of H. pylori isolates of our investigation were resistant to at least one antimicrobial agents. In keeping with this, 12.5 % of isolates were resistant to more than 6 antimicrobial agents. In addition, the highest levels of resistance were seen against six types of antimicrobial agents. These antimicrobial agents are the best choices for treatment of the cases of infection with H. pylori. In fact, in most cases of peptic ulcer diseases with the sign of H. pylori infection, these antimicrobial agents are used. Unfortunately, prescription of these types of antibiotics are usually irregular and over than allowance levels. Therefore, it is not surprising that majority of H. pylori strains were resistant to these antimicrobial agents. Similar results have been described by Bang et al. (2007) [29], Thyagarajan et al. (2003) [30], Yahaghi et al. (2014) [13] and Secka et al. (2013) [31]. High resistance levels of H. pylori strains of biopsy samples taken from patients with peptic ulcer diseases against metronidazole (34.7 %), clarithromycin (16.7 %) and amoxicillin (11.8 %) was reported previously by Bang et al. (2007) [29]. Another Iranian investigation [32] showed that the prevalence of resistance in H. pylori strains of gastrointestinal disorders against metronidazole, clarithromycin and amoxicillin had the ranges of 4 to 57 %. Bahrami et al. (2011) [33] reported that the high prevalence of water-borne H. pylori resistance against furazolidone (4.5 %), clarithromycin (0.9 %), amoxicillin (0 %), metronidazole (36.4 %) and tetracycline (1.8 %). Considerable levels of resistance in H. pylori strains of clinical specimens against metronidazole, clarithromycin, quinolones, amoxicillin, and tetracycline have been reported from various countries such as Nigeria, Senegal, India, Saudi-Arabia, China, Colombia, Taiwan, Brazil, Thailand, Argentina and Egypt [34].
The bottled mineral water samples of our study harbored H. pylori positive in genotypes of vacA, cagA, cagE, iceA, oipA and babA2. The most commonly detected genotypes were vacAs1a (100 %), vacAm1a (87.5 %), cagA (62.5 %) and iceA1 (62.5 %). Low prevalence of oipA (25 %), babA2 (25 %) and cagE (37.5 %) genotypes was also detected. The number of isolates which were positive for combined genotypes was also considerable. M1as1a (62.5 %), m1as2 (37.5 %) and m2s2 (37.5 %) and also s1a/cagA+/IceA2/oipA-/babA2-/cagE- (50 %) were the most commonly detected combined genotypes. As it show, cagA, oipA, babA2 and cagE negative strains of H. pylori were more prevalent than their positive strains. The bottled mineral water samples weren’t harbored the cagA, cagE, babA2 and oipa positive strains of H. pylori. The number of iceA2 strains was also low. In keeping with the uncertain cause of low distribution of cagA, oipA, babA2 and cagE genotypes in the H. pylori strains of bottled mineral water, high prevalence of pathogenic genotypes can guarantee the occurrence of gastrointestinal disorders due to consumption of H. pylori positive bottled mineral water samples of our study. The number of studies which were conducted on genotyping of H. pylori isolated from bottled mineral water are significantly low. In an only study which was conducted by Yingzhi et al. (2002) [35], high presence of vacAs1a, vacAm1a and vacAs1am1a genotypes in the drinking water samples was reported. There were no additional previously published data in this field on drinking water but high presence of vacAs1a, vacAm1a, vacAs2 alleles and also m1as1a, m1as2, m1as1b, m1bs1b, m2s1a, m2s2 and m1am2 genotypes in the H. pylori strains of human clinical samples such as gastric biopsy, feces and saliva have been reported previously [36–38]. Sedaghat et al. (2014) [39] reported that the prevalence of cagA, iceA1, iceA2, oipA and babA2 genotypes in the cases of human clinical disorders including gastric ulcer and gastritis were 62.2, 48.6, 16.2, 81.1 and 94.6 %, respectively which was similar to our finding in bottled mineral water. High potential of vacA positive strains of H. pylori for causing human clinical disorders has been reported previously [11–13, 40]. The severity of diseases caused by babA positive strains of H. pylori is higher than babA negative strains [41]. We found that 25 % of H. pylori strains recovered from the bottled mineral water samples harbored the babA genotype. The expression of iceA1 is up-regulated on contact between H. pylori and human epithelial cells and may be related with peptic ulcer disease. We found that 62.5 % of our strains were iceA1 +. The expression of the oipA associated with IL-8 induction and is related with severe clinical outcomes [11–13, 40]. Prevalence of this genotype in the H. pylori strains of our investigation were 25 %.
Conclusions
To our best knowledge, the present investigation is the first prevalence report for presence of H. pylori in bottled mineral water as well as its genotyping and antimicrobial resistance properties all-around the world. Bottled mineral water samples of Iranian factories harbored virulent and resistant strains of H. pylori which shows an important public health problem. Brand C, July month, resistance against ampicillin, s1a allele, m1as1a genotype and s1a/cagA+/IceA2/oipA-/babA2-/cagE combined genotypes were the most commonly detected characters of H. pylori isolates of bottled mineral water samples of our study. Cefsulodin and furazolidone prescription can be effective for treatment of cases of H. pylori.
Methods
Sample collection
According to the prevalence of H. pylori in water samples of some previous studies, the number of samples was calculated based on the following formula:
From April 2014 to April 2015, overall 450 bottled mineral water samples of various brands (A-D) were randomly collected from the Isfahan province, Iran. Bottled mineral water samples were collected based on the protocols presented by International Standard Organization (ISO 5667-1:1980, ISO 5667-2:1991 and ISO 5667-4:1987). Samples (100 mL) were transported to the lab on ice, and used within 2 h of collection.
Isolation of Helicobacter pylori
Samples were filtered through 0.045 μm filter membranes (Albet Co.). Each membrane was then immersed into 2 mL of Tryptic Soy Broth (TSB, Merck) for 1 h. After that, each 2 mL TSB was taken and cultured for H. pylori. Samples were cultured on Brucella agar (Merck, Germany) containing campylobacter selective supplement (5 mg/L, Merck), trimethoprim (0.25 mg/L), colistin methanesulfonate (30 mg/L), cycloheximide (100 mg/L), nalidixic acid (30 mg/L), trimethoprim (30 mg/L), vancomycin (10 mg/L) (Sigma, St. Louis, MO, USA), amphotericin B, sheep blood (5 %), and 7 % fetal calf serum (Sigma). After 72 h incubation at 37 °C in microaerophilic condition (85 % N2, 10 % CO2 and 5 % O2) using MART system (Anoxamat, Lichtenvoorde, The Netherlands), the bacterial growth was tested and confirmed as H. pylori using the gram staining, urease, and oxidase tests.
Antimicrobial susceptibility testing
Pure cultures of H. pylori were applied for antibiotic susceptibility test. One strain from each H. pylori-positive sample was selected for this aim. Antimicrobial susceptibility test was accomplished by the Kirby-Bauer disc diffusion method using Mueller-Hinton agar (Merck, Germany) supplemented with 5 % defibrinated sheep blood and 7 % fetal calf serum, according to the Clinical Laboratory Standards Institute (CLSI 2012) [42]. The antimicrobial resistance of H. pylori was measured against the widely used antibiotics in cases of H. pylori gastric ulcer. The following antimicrobial disks (HiMedia Laboratories, Mumbai, India) were used: ampicillin (10 μg), metronidazole (5 μg), erythromycin (5 μg), clarithromycin (2 μg), amoxicillin (10 μg), levofloxacin (5 μg), cefsulodin (30 μg), trimethoprim (25 μg), furazolidone (1 μg), moxifloxacin (5 μg), tinidazole (prepared from pure powders, Sigma, 5 μg) and ciprofloxacin (5 μg). After incubation at 37 °C for 48 h in a microaerophilic atmosphere (85 % N2, 10 % CO2 and 5 % O2,), the susceptibility of the H. pylori was measured against each antimicrobial agents. Results were construed in accordance with interpretive criteria provided by CLSI (2012) [42]. The H. pylori ATCC 43504 was used a quality control organism in antimicrobial susceptibility determination.
DNA extraction and Helicobacter pylori 16S rRNA gene amplification
Suspected colonies were also identified as H. pylori based on the PCR technique. Genomic DNA was extracted from the colonies with typical characters of H. pylori using a DNA extraction kit for cells and tissues (Roche Applied Science, Germany, 11814770001) according to the manufacturer’s instructions and its density was assessed by optic densitometry. Extracted DNA was amplified for the 16S rRNA gene (primers: HP-F: 5'-CTGGAGAGACTAAGCCCTCC-3' and HP-R: 5'-ATTACTGACGCTGATTGTGC-3') [43]. PCR reactions were performed in a final volume of 50 μL containing 5 μL 10 × buffer + MgCl2, 2 mM dNTP, 2 unit Taq DNA polymerase, 100 ng genomic DNA as a template, and 25 picomole of each primer. PCR was performed using a thermal cycler (Eppendorf Co., Germany) under the following conditions: an initial denaturation for 2 min at 94 °C; 30 cycles of 95 °C for 30 s, 60 °C for 30 s, and 72 °C for 30 s and a final extension at 72 °C for 8 min.
Genotyping of vacA, cagA, cagE, iceA, babA2 and oipA genotypes in the Helicobacter pylori isolates of drinking water
Presence of the iceA1, iceA2. oipA, cagA, cagE, babA2 genotypes and also various genotypes of vacA alleles (s1a, s1b, s1c, m1a, m1b and m2) were determined using PCR technique. List of primers and PCR program are shown in Table 6 [44–51]. PCR amplifications were performed in a programmable thermal cycler (Master Cycle Gradiant, Eppendorf, Germany) and all runs included one negative DNA control consisting of PCR grade water and two or more positive controls (26695, J99, SS1, Tx30, 88–23 and 84–183).
Table 6.
Oligonucleotide primers and PCR conditions used for genotyping of Helicobacter pylori strains isolated from Iranian bottled mineral water
| Genes | Primer Sequence (5’-3’) | Size of product (bp) | Volume of PCR reaction (50 μl) | PCR programs |
|---|---|---|---|---|
| vacA s 1 a | F: CTCTCGCTTTAGTAGGAGC | 213 | 5 μL PCR buffer 10X | 1 cycle: 95 °C ------------ 1 min. |
| R: CTGCTTGAATGCGCCAAAC | ||||
| vacA s 1 b | F: AGCGCCATACCGCAAGAG | 187 | 1.5 mM Mgcl2 | 32 cycle: 95 °C ------------ 45 s |
| CTGCTTGAATGCGCCAAAC | 200 μM dNTP (Fermentas) | |||
| vacA s 1 c | F: CTCTCGCTTTAGTGGGGYT | 213 | 64 °C ------------ 50 s | |
| R: CTGCTTGAATGCGCCAAAC | 0.5 μM of each primers F & R | |||
| vacA s 2 | F: GCTAACACGCCAAATGATCC | 199 | 72 °C ------------ 70 s | |
| R: CTGCTTGAATGCGCCAAAC | 1.25 U Taq DNA polymerase (Fermentas) | |||
| vacA m 1 A | F: GGTCAAAATGCGGTCATGG | 290 | 1 cycle: 72 °C ------------ 5 min | |
| R: CCATTGGTACCTGTAGAAAC | ||||
| vacA m 1 B | F: GGCCCCAATGCAGTCATGGA | 291 | 2.5 μL DNA template | |
| R: GCTGTTAGTGCCTAAAGAAGCAT | ||||
| vacA m 2 | F: GGAGCCCCAGGAAACATTG | 352 | ||
| R: CATAACTAGCGCCTTGCA | ||||
| cag A | F: GATAACAGCCAAGCTTTTGAGG | 300 | 5 μL PCR buffer 10X | 1 cycle: 94 °C ------------ 1 min. |
| R: CTGCAAAAGATTGTTTGGCAGA | ||||
| 2 mM Mgcl2 | 32 cycle: 95 °C ------------ 60 s | |||
| 150 μM dNTP (Fermentas) | ||||
| 0.75 μM of each primers F & R | 56 °C ------------ 60 s | |||
| 1.5 U Taq DNA polymerase (Fermentas) | 72 °C ------------ 60 s | |||
| 1 cycle: 72 °C ------------ 10 min | ||||
| 3 μL DNA template | ||||
| iceA 1 | F: GTGTTTTTAACCAAAGTATC | 247 | 5 μL PCR buffer 10X | 1 cycle: 94 °C ------------ 1 min. |
| R: CTATAGCCASTYTCTTTGCA | ||||
| iceA 2 | F: GTTGGGTATATCACAATTTAT | 229/334 | 2 mM Mgcl2 | 32 cycle: 94 °C ------------ 60 s |
| R: TTRCCCTATTTTCTAGTAGGT | 200 μM dNTP (Fermentas) | |||
| 56 °C ------------ 60 s | ||||
| 0.5 μM of each primers F & R | ||||
| 72 °C ------------ 60 s | ||||
| 1.5 U Taq DNA polymerase (Fermentas) | ||||
| 1 cycle: 72 °C ------------ 8 min | ||||
| 5 μL DNA template | ||||
| oip A | F: GTTTTTGATGCATGGGATTT | 401 | 5 μL PCR buffer 10X | 1 cycle: 94 °C ------------ 2 min. |
| R: GTGCATCTCTTATGGCTTT | ||||
| 2.5 mM Mgcl2 | 32 cycle: 94 °C ------------ 60 s | |||
| 200 μM dNTP (Fermentas) | ||||
| 56 °C ------------ 60 s | ||||
| 0.5 μM of each primers F & R | ||||
| 72 °C ------------ 60 s | ||||
| 2 U Taq DNA polymerase (Fermentas) | ||||
| 1 cycle: 72 °C ------------ 10 min | ||||
| 3 μL DNA template | ||||
| cagE | F: TTGAAAACTTCAAGGATAGGATAGAGC R: GCCTAGCGTAATATCACCATTACCC | 508 | 5 μL PCR buffer 10X | 1 cycle: 94 °C ------------ 1 min. |
| 2 mM Mgcl2 | 35 cycle: 94 °C ------------ 60 s | |||
| 150 μM dNTP (Fermentas) | ||||
| 53 °C ------------ 45 s | ||||
| 0.75 μM of each primers F & R | ||||
| 72 °C ------------ 45 min | ||||
| 1.5 U Taq DNA polymerase (Fermentas) | ||||
| 1 cycle: 72 °C ------------ 8 min | ||||
| 3 μL DNA template | ||||
| BabA2 | F: CCAAACGAAACAAAAAGCGT | 271 | 5 μL PCR buffer 10X | 1 cycle: 95 °C ------------ 1 min. |
| R: GCTTGTGTAAAAGCCGTCGT | ||||
| 1.5 mM Mgcl2 | 30 cycle: 91 °C ------------ 60 s | |||
| 200 μM dNTP (Fermentas) | ||||
| 45 °C ------------ 60 s | ||||
| 0.5 μM of each primers F & R | ||||
| 72 °C ------------ 60 s | ||||
| 1.25 U Taq DNA polymerase (Fermentas) | ||||
| 1 cycle: 72 °C ------------ 8 min | ||||
| 2.5 μL DNA template |
Gel electrophoresis
The PCR amplification products (10 μl) were subjected to electrophoresis in a 2.5 % agarose gel in 1X TBE buffer at 80 V for 30 min, stained with ethidium bromide, and images were obtained in a UVIdoc gel documentation systems (UK). The PCR products were identified by 100 bp DNA size marker (Fermentas, Germany) [52].
Statistical analysis
Data were transferred to Microsoft Excel spreadsheets (Microsoft Corp., Redmond, WA, USA) for analysis. Using SPSS 16.0 statistical software (SPSS Inc., Chicago, IL, USA), Chi–square test and Fisher’s exact two-tailed test analysis was performed and differences were considered significant at values of P <0.05. Distribution of H. pylori genotypes isolated from bottled mineral water were statistically analyzed.
Abbreviations
- AM
Ampicillin
- AMX
Amoxicillin
- babA
Blood group antigen-binding adhesion
- cag
Cytotoxin associated gene
- Cef
Cefsulodin
- CLR
Clarithromycin
- CLSI
Clinical Laboratory Standards Institute
- ER
Erythromycin
- FZL
Furazolidone
- H. pylori
Helicobacter pylori
- iceA
Induced by contact with the epithelium antigen
- Lev
Levofloxacin
- Met
Metronidazole
- oip
Outer inflammatory protein
- PCR
Polymerase chain reaction
- TRP
Trimethoprim
- vacA
Vacuolating cytotoxin
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
RR, FK, NJ-J and ER contributed equally to this work. All authors read and approved the final manuscript.
Contributor Information
Reza Ranjbar, Email: Ranjbarre@gmail.com.
Faham Khamesipour, Email: Dr_Faham@yahoo.com.
Nematollah Jonaidi-Jafari, Email: Jonaidi2000@yahoo.com.
Ebrahim Rahimi, Email: Ebrahimrahimi55@yahoo.com.
References
- 1.Bahrami AR, Rahimi E, Ghasemian SH. Detection of Helicobacter pylori in City Water, Dental Units’ Water, and Bottled Mineral Water in Isfahan. Iran. Sci World J. 2013;2013:280510. doi: 10.1155/2013/280510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Momtaz H, Dehkordi FS, Rahimi E, Asgarifar A. Detection of Escherichia coli, Salmonella species, and Vibrio cholerae in tap water and bottled drinking water in Isfahan, Iran. BMC Public Health. 2013;7(13):556. doi: 10.1186/1471-2458-13-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shrestha S, Paudel P, Pradhan GB, Shrestha L, Bhattachan CL. Prevalence study of H. pylori infection in dyspeptic patients coming to Nepal Medical College Teaching Hospital, Jorpati, Kathmandu. Nepal Med Coll J. 2012;14:229–233. [PubMed] [Google Scholar]
- 4.Suzuki R, Shiota S, Yamaoka Y. Molecular epidemiology, population genetics, and pathogenic role of Helicobacter pylori. Infect Genet Evol. 2012;12:203–213. doi: 10.1016/j.meegid.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vu C, Ng YY. Prevalence of Helicobacter pylori in peptic ulcer disease in a Singapore hospital. Singapore Med J. 2000;41:478–481. [PubMed] [Google Scholar]
- 6.Mastromarino P, Conti C, Donato K, et al. Does hospital work constitute a risk factor for Helicobacter pylori infection. J Hosp Infect. 2005;60:261–268. doi: 10.1016/j.jhin.2004.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown LM. Helicobacter pylori: epidemiology and routes of transmission. Epidemiol Rev. 2000;22(2):283–297. doi: 10.1093/oxfordjournals.epirev.a018040. [DOI] [PubMed] [Google Scholar]
- 8.Percival SL, Thomas JG. Transmission of Helicobacter pylori and the role of water and biofilms. J Water Health. 2009;7(3):469–477. doi: 10.2166/wh.2009.070. [DOI] [PubMed] [Google Scholar]
- 9.El-Sharouny E, El-Shazli H, Olama Z. Detection of Helicobacter pylori DNA in Some Egyptian Water Systems and Its Incidence of Transmission to Individuals. Iran J Public Health. 2015;44(2):203–210. [PMC free article] [PubMed] [Google Scholar]
- 10.Arévalo-Galvis A, Trespalacios-Rangell AA, Otero W, Mercado-Reyes MM, Poutou-Piñales RA. Prevalence of cagA, vacA, babA2 and iceA genes in H. pylori strains isolated from Colombian patients with functional dyspepsia. Pol J Microbiol. 2012;61(1):33–40. [PubMed] [Google Scholar]
- 11.Momtaz H, Dabiri H, Souod N, Gholami M. Study of Helicobacter pylori genotype status in cows, sheep, goats and human beings. BMC Gastroenterol. 2014;14:61. doi: 10.1186/1471-230X-14-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mousavi S, Safarpoor Dehkordi F, Rahimi E. Virulence factors and antibiotic resistance of Helicobacter pylori isolated from raw milk and unpasteurized dairy products in Iran. J Venom Anim Toxins Incl Trop Dis. 2014;20:51. doi: 10.1186/1678-9199-20-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yahaghi E, Khamesipour F, Mashayekhi F, Safarpoor Dehkordi F, Sakhaei MH, Masoudimanesh M, et al. Helicobacter pylori in vegetables and salads: genotyping and antimicrobial resistance properties. Biomed Res Int. 2014;2014:757941. doi: 10.1155/2014/757941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramis IB, Vianna JS, Silva Junior LV, Von Groll A, Silva PE. cagE as a biomarker of the pathogenicity of Helicobacter pylori. Rev Soc Bras Med Trop. 2013;46(2):185–189. doi: 10.1590/0037-8682-0054-2012. [DOI] [PubMed] [Google Scholar]
- 15.Markovska R, Boyanova L, Yordanov D, Gergova G, Mitov I. Helicobacter pylori oipA genetic diversity and its associations with both disease and cagA, vacA s, m, and i alleles among Bulgarian patients. Diagn Microbiol Infect Dis. 2011;71(4):335–340. doi: 10.1016/j.diagmicrobio.2011.08.008. [DOI] [PubMed] [Google Scholar]
- 16.Siavoshi F, Malekzadeh R, Daneshmand M, Ashktorab H. Helicobacter pylori endemic and gastric disease. Dig Dis Sci. 2005;50(11):2075–2080. doi: 10.1007/s10620-005-3010-1. [DOI] [PubMed] [Google Scholar]
- 17.Banatvala N, Davies GR, Abdi Y, Clements L, Rampton DS, Hardie JM, et al. High prevalence of Helicobacter pylori metronidazole resistance in migrants to east London: relation with previous nitroimidazole exposure and gastroduodenal disease. Gut. 1994;35(11):1562–1566. doi: 10.1136/gut.35.11.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuipers EJ, Meijer GA. Helicobacter pylori gastritis in Africa. Eur J Gastroenterol Hepatol. 2000;12(6):601–603. doi: 10.1097/00042737-200012060-00003. [DOI] [PubMed] [Google Scholar]
- 19.Shi R, Xu S, Zhang H, Ding Y, Sun G, Huang X, et al. Prevalence and risk factors for Helicobacter pylori infection in Chinese populations. Helicobacter. 2008;13(2):157–165. doi: 10.1111/j.1523-5378.2008.00586.x. [DOI] [PubMed] [Google Scholar]
- 20.Vilaichone RK, Mahachai V, Shiota S, Uchida T, Ratanachu-ek T, Tshering L, et al. Extremely high prevalence of Helicobacter pylori infection in Bhutan. World J Gastroenterol. 2013;19(18):2806–2810. doi: 10.3748/wjg.v19.i18.2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Watson CL, Owen RJ, Said B, Lai S, Lee JV, Surman-Lee S, et al. Detection of Helicobacter pylori by PCR but not culture in water and biofilm samples from drinking water distribution systems in England. J Appl Microbiol. 2004;97(4):690–698. doi: 10.1111/j.1365-2672.2004.02360.x. [DOI] [PubMed] [Google Scholar]
- 22.Nurgalieva ZZ, Malaty HM, Graham DY, Almuchambetova R, Machmudova A, Kapsultanova D, et al. Helicobacter pylori infection in Kazakhstan: effect of water source and household hygiene. Am J Trop Med Hyg. 2002;67:201–206. doi: 10.4269/ajtmh.2002.67.201. [DOI] [PubMed] [Google Scholar]
- 23.Gião MS, Azevedo NF, Wilks SA, Vieira MJ, Keevil CW. Persistence of Helicobacter pylori in Heterotrophic Drinking-Water Biofilms. Appl Environ Microbiol. 2008;74(19):5898–5904. doi: 10.1128/AEM.00827-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Azevedo NF, Pinto AR, Reis NM, Vieira MJ, Keevil CW. Shear stress, temperature, and inoculation concentration influence the adhesion of water-stressed Helicobacter pylori to stainless steel 304 and polypropylene. Appl Environ Microbiol. 2006;72:2936–2941. doi: 10.1128/AEM.72.4.2936-2941.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hegarty JP, Dowd MT, Baker KH. Occurrence of Helicobacter pylori in surface water in the United States. J Appl Microbiol. 1999;87:697–701. doi: 10.1046/j.1365-2672.1999.00912.x. [DOI] [PubMed] [Google Scholar]
- 26.Flanigan D, Rodgers M. A Method to detect viable Helicobacter pylori bacteria in groundwater. Acta Hydrochim Hydrobiol. 2003;31:45–48. doi: 10.1002/aheh.200390015. [DOI] [Google Scholar]
- 27.Nayak AK, Rose JB. Detection of Helicobacter pylori in sewage and water using a new quantitative PCR method with SYBR green. J Appl Microbiol. 2007;103(5):1931–1941. doi: 10.1111/j.1365-2672.2007.03435.x. [DOI] [PubMed] [Google Scholar]
- 28.Konishi K, Saito N, Shoji E, Takeda H, Kato M, Asaka M, et al. Helicobacter pylori: longer survival in deep ground water and sea water than in a nutrient-rich environment. APMIS. 2007;115:1285–1291. doi: 10.1111/j.1600-0643.2007.00594.x. [DOI] [PubMed] [Google Scholar]
- 29.Bang SY, Han DS, Eun CS, Kim JE, Ahn SB, Sohn JH, et al. Changing patterns of antibiotic resistance of Helicobacter pylori in patients with peptic ulcer disease. Korean J Gastroenterol. 2007;50:356–362. [PubMed] [Google Scholar]
- 30.Thyagarajan SP, Ray P, Das BK, Ayyagari A, Khan AA, Dharmalingam S, et al. Geographical difference in antimicrobial resistance pattern of Helicobacter pylori clinical isolates from Indian patients: Multicentric study. J Gastroenterol Hepatol. 2003;18:1373–1378. doi: 10.1046/j.1440-1746.2003.03174.x. [DOI] [PubMed] [Google Scholar]
- 31.Secka O, Berg DE, Antonio M, Corrah T, Tapgun M, Walton R, et al. Antimicrobial susceptibility and resistance patterns among Helicobacter pylori strains from The Gambia, West Africa. Antimicrob Agents Chemother. 2013;57:1231–1237. doi: 10.1128/AAC.00517-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mirzaei N, Poursina F, Faghri J, Talebi M, Khataminezhad MR, Hasanzadeh A, et al. Prevalence of resistance to Helicobacter pylori strains to selected antibiotics in Isfahan, Iran. J J Microbiol. 2013;6:e6342. [Google Scholar]
- 33.Bahrami AR, Aminipour Harandi MR, Kazemi Kheirabadi E, Sharifian B, Ghasemian Safaei H, Rahimi E. Antimicrobial Resistance of Helicobacter pylori Isolates from Cow Feces, Raw Milk and Drinking Water in Iran, Middle East. J Sci Res. 2011;10(6):698–701. [Google Scholar]
- 34.World Gastroenterology Organization (WGO) Global Guideline . Helicobacter pylori in developing countries. 2010. [Google Scholar]
- 35.Lu Y, Redlinger TE, Avitia R, Galindo A, Goodman K. Isolation and Genotyping of Helicobacter pylori from Untreated Municipal Wastewater. Appl Environ Microbiol. 2002;68:1436–1439. doi: 10.1128/AEM.68.3.1436-1439.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saribasak H, Salih BA, Yamaoka Y, Sander E. Analysis of Helicobacter pylori genotypes and correlation with clinical outcome in Turkey. J Clin Microbiol. 2004;42(4):1648–1651. doi: 10.1128/JCM.42.4.1648-1651.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miernyk K, Morris J, Bruden D, McMahon B, Hurlburt D, Sacco F, et al. Characterization of Helicobacter pylori cagA and vacAGenotypes among Alaskans and their correlation with clinical disease. J Clin Microbiol. 2011;49(9):3114–3121. doi: 10.1128/JCM.00469-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Havaei SA, Mohajeri P, Khashei R, Salehi R, Tavakoli H. Prevalence of Helicobacter pylori vacA different genotypes in Isfahan, Iran. Adv Biomed Res. 2014;3:48. doi: 10.4103/2277-9175.125761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sedaghat H, Moniri R, Jamali R, Arj A, Razavi Zadeh M, Moosavi SGA, et al. Prevalence of Helicobacter pylori vacA, cagA, cagE, iceA, babA2, and oipA genotypes in patients with upper gastrointestinal diseases. Iran J Microbiol. 2014;6(1):14–21. [PMC free article] [PubMed] [Google Scholar]
- 40.Torres LE, Melián K, Moreno A, et al. Prevalence of vacA, cagA and babA2 genes in Cuban Helicobacter pylori isolates. World J Gastroenterol. 2009;15:204–210. doi: 10.3748/wjg.15.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ben Mansour K, Fendri C, Zribi M, Masmoudi A, Labbene M, Fillali A, et al. Prevalence of Helicobacter pylori vacA, cagA, iceA and oipA genotypes in Tunisian patients. Ann Clin Microbiol Antimicrob. 2010;19(9):1. doi: 10.1186/1476-0711-9-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clinical and Laboratory Standards Institute (CLSI). Performance standards for antimicrobial susceptibility testing. Twenty-second informational supplement, M100-S22. Clinical and Laboratory Standards Institute, Wayne, PA, 2012.
- 43.Ho SA, Hoyle JA, Lewis FA. Direct polymerase chain reaction test for detection of Helicobacter pylori in humans and animals. J Clin Microbiol. 1991;29:2543–2549. doi: 10.1128/jcm.29.11.2543-2549.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Versalovic J, Koeuth T, Lupski JR. Distribution of repetitive DNA sequences in Eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991;19:6823–6831. doi: 10.1093/nar/19.24.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tumurru MK, Cover TL, Blaser MJ. Cloning and expression of a highmolecular mass major antigen of Helicobacter pylori: evidence of linkage to cytotoxin production. Infect Immun. 1993;61:1799–1809. doi: 10.1128/iai.61.5.1799-1809.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peek RM, Thompson SA, Donahue JP, et al. Adherence to gastric epithelial cells induces expression of a Helicobacter pylori gene, iceA, that is associated with clinical outcome. Proc Assoc Am Physicians. 1998;110:531–544. [PubMed] [Google Scholar]
- 47.Wang J, Chi DS, Laffan JJ, Li C, Ferguson DA, Jr, Litchfield P, et al. Comparison of cytotoxin genotypes of Helicobacter pylori in stomach and saliva. Dig Dis Sci. 2002;47:1850–1856. doi: 10.1023/A:1016417200611. [DOI] [PubMed] [Google Scholar]
- 48.Kauser F, Hussain MA, Ahmed I, Habeeb A, Khan AA, Ahmed N. Comparing genomes of Helicobacter pylori strains from the high-altitude desert of Ladakh, India. J Clin Microbiol. 2005;43:1538–1545. doi: 10.1128/JCM.43.4.1538-1545.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamazaki S, Yamakawa A, Okuda T, Ohtani M, Suto H, Ito Y, et al. Distinct diversity of vacA, cagA, and cagE genes of Helicobacter pylori associated with peptic ulcer in Japan. J Clin Microbiol. 2005;43:3906–3916. doi: 10.1128/JCM.43.8.3906-3916.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chomvarin C, Namwat W, Chaicumpar K, Mairiang P, Sangchan A, Sripa B, et al. Prevalence of Helicobacter pylori vacA, cagA, cagE, iceA and babA2 genotypes in Thai dyspeptic patients. Int J Infect Dis. 2008;12:30–36. doi: 10.1016/j.ijid.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 51.Thirumurthi S, Graham DY. Helicobacter pylori infection in India from a western perspective. Ind J Med Res. 2012;136:549–562. [PMC free article] [PubMed] [Google Scholar]
- 52.Ling CL, Joss AW, Davidson MM, Ho-Yen DO. Identification of different Borrelia burgdorferi genomic groups from Scottish ticks. Mol Pathol. 2000;53(2):94–8. doi: 10.1136/mp.53.2.94. [DOI] [PMC free article] [PubMed] [Google Scholar]


