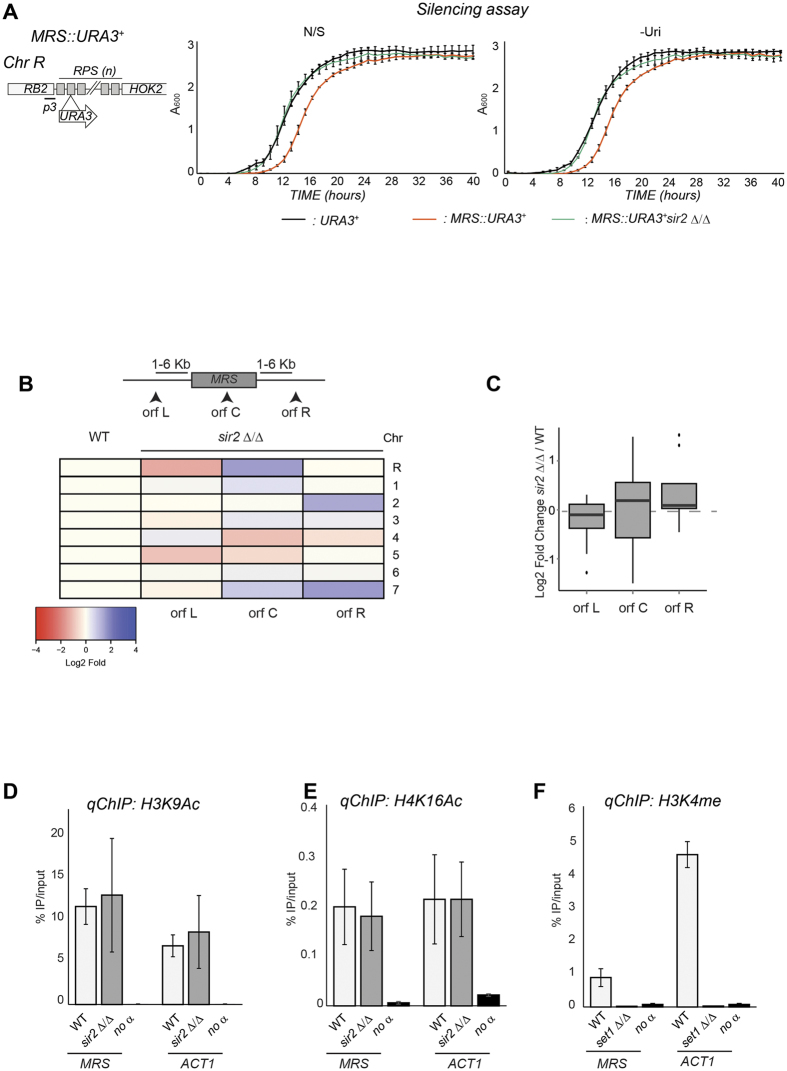
Figure 3. MRS repeats are assembled into transcriptionally-permissive chromatin.
(A) Top panel: Schematic of MRS::URA3+ reporter strain. Bottom panel: Silencing assay of the MRS::URA3+ reporter strain in WT and sir2 Δ/Δ isolates. A URA+ (URA3+) strain was included as a control. (B,C) RNA deep-sequencing of sir2 Δ/Δ and WT isolates. (B) Normalised read counts (FPKM) of MRS associated (MRS-C) and proximal (MRS-L and MRS-R) genes were calculated from RNA-seq data for WT and sir2 Δ/Δ isolates. The heat-map depicts the log2 fold ratio of FPKM data between sir2 Δ/Δ and WT isolates. (C) Boxplot showing log2 fold changes in transcriptional expression for MRS-internal (MRS-C) and adjacent (MRS-L and MRS-R) genes between sir2 Δ/Δ and WT isolates. (D,E) qChIP to detect H3K9Ac, H4K16Ac levels associated with the MRS repeats and ACT1 in WT and sir2 Δ/Δ isolates and (F) H3K4me2 levels associated with the MRS repeats and ACT1 in WT and set1 Δ/Δ isolates. Error bars in each panel: Standard deviation (SD) of three biological replicates.