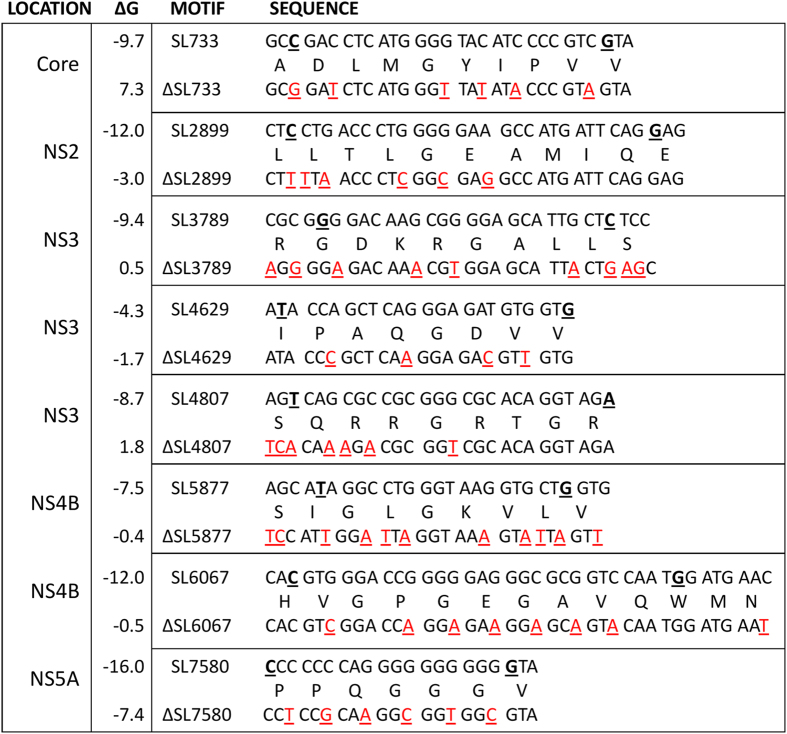
Figure 5. Silent mutations designed to disrupt the secondary structures of the pPSs within JFH-1.
Multiple silent mutations (red, underlined) were incorporated into the pJFH-1 plasmid to disrupt the pPSs. The 5′ and 3′ nucleotides of the structures displayed from Fig. 4 are underlined in bold in the wildtype (upper) sequences; on occasion additional nucleotides immediately flanking these regions were mutated to maximise the disruption potential. Mutant genomes were named according to the disrupted stem-loop (ΔSL). The peptides encoded within each motif are described between the nucleotide sequences. ΔG refers to the free energy of the most stable predicted RNA secondary structure (kcal/mol) as calculated by mFold21.