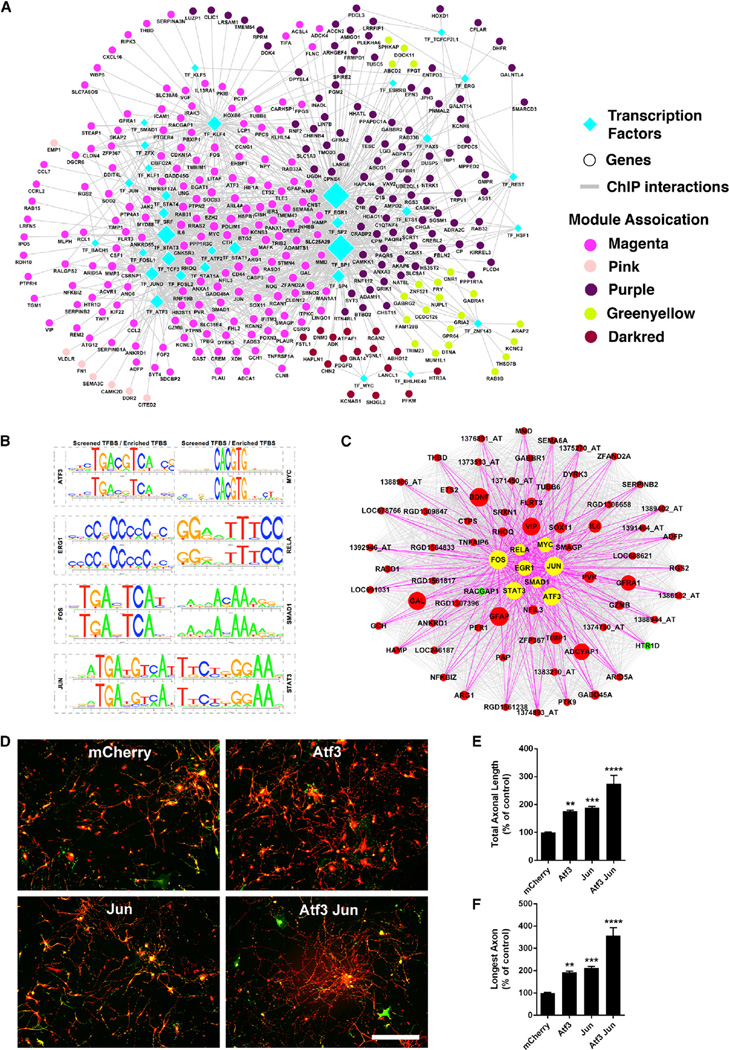
Figure 3. TF-Binding Site Enrichment in Injured Regenerating Neurons.
(A) Regulatory network of differentially expressed genes after nerve injury. Nodes correspond to genes or TFs and edges to ChIP interaction. Node color represents their corresponding module associations as denoted in the legends and over-represented TFs (diamond) are shown in cyan. Node size is based on its centrality.
(B) Sequence logo plots of reference (JASPAR/TRANSFAC) and identified position weight matrix for each TF significantly over-represented in magenta module are shown.
(C) Magenta module, showing eight over-represented TFs as hub genes. Nodes correspond to genes and edges to significant correlation. Larger nodes correspond to number of PubMed hits with co-occurrence of gene and neuronal regeneration, axonal regeneration, and nerve injury tags; upregulated (red), downregulated (green), and over-represented TFs (yellow) are shown. Only edges connected by over-represented TFs are highlighted.
(D–F) Combined TF overexpression. (D) Photomicrographs of dissociated DRG neurons transduced with the indicated viruses at 1 DIV, replated at 7 DIV, and cultured for an additional 20 hr on laminin. Red, bIII-tubulin; green, EGFP. (E and F) Quantification of total axonal length (E) and longest axon (F) reveal significant enhancement of axonal growth for ATF3 and JUN expression individually, compared to control (mCherry). Combined ATF3 and JUN overexpression enhances axonal growth significantly more than ATF3 or JUN individually. Data are mean ± SE. n = 30 wells. **p < 0.01, ***p < 0.001, ****p < 0.0001, one-way ANOVA with Bonferroni’s post hoc test. Scale bar, 200 microns.