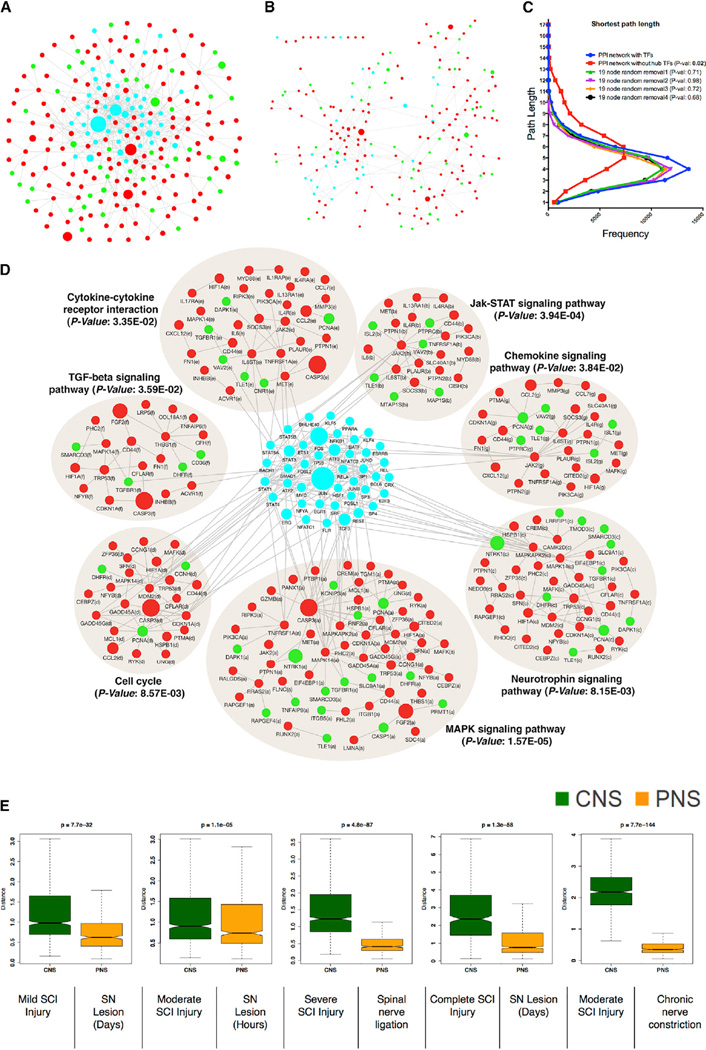
Figure 4. Over-Represented TFs Are Involved in Transcriptional Cross-Talk between Regeneration-Associated Pathways.
(A) Protein-protein interaction (PPI) network of differentially expressed genes after nerve injury. Nodes correspond to genes and edges to PPI. Larger nodes correspond to number of PubMed hits with co-occurrence of gene and neuronal regeneration, axonal regeneration, and nerve injury tags. Node color represents upregulated (red), downregulated (green), and over-represented (cyan) TFs.
(B) PPI network dissociation after in silico removal of 19 hub TFs is shown.
(C) Distribution of the shortest path between pairs of nodes in the PPI network with or without in silico removal of 19 hub TFs. Random removal of a similar number of nodes is shown for comparison.
(D) Significantly enriched KEGG pathways (Benjamini-corrected p values < 0.05) in the PPI network.
(E) Boxplot representation of the variability in the expression levels of the over-represented TFs between CNS and PNS injuries (see Figures S1C and S1D). Time series data after CNS or PNS injury (see Figure S2) were used to create distance matrix using Euclidean distance measure to create the boxplot. Non-parametric Kruskal-Wallis test was used to compare differences between CNS and PNS injury datasets.