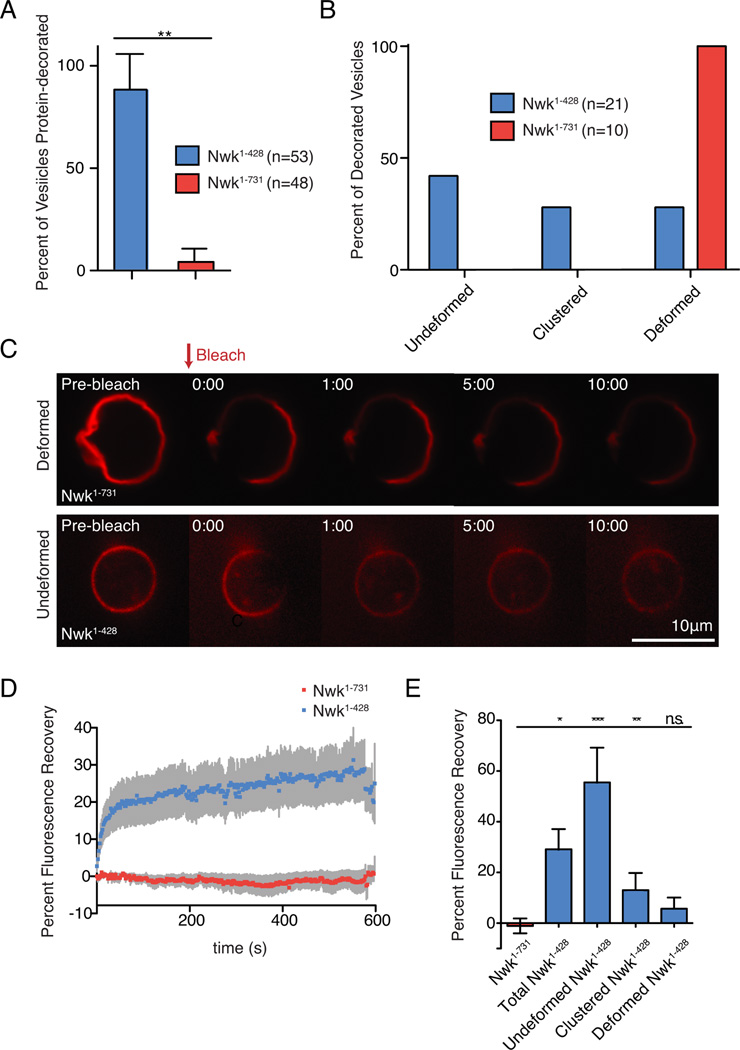
Figure 5. Nwk SH3 domains limit promiscuous membrane binding, and promote stable F-BAR protein scaffolds associated with membrane remodeling.
NBD-PE-labeled GUVs incubated with SNAP-549-tagged Nwk variants (500 nM) were imaged by spinning disk confocal microscopy. Lipid composition was [DOPC:POPE:DOPS:PI(4,5)P2:NBD-PE] = 75:14.5:5:5:.5. n represents the number of vesicles examined. (A) Percent of protein-decorated GUVs after 30-minute incubation with Nwk1–428 (blue) or Nwk1–731 (red). Graph represents mean +/− s.e.m. from 3 independent experiments. Data are identical to 5% PI(4,5)P2 data point in Figs. 6B, C. (B) Quantification of the morphology of protein-decorated GUVs after 30-minute incubation with purified Nwk1–428 or Nwk1–731. GUVs were imaged from n independent reactions. (C) Single spinning disc confocal slices of GUVs showing Nwk1–731 displaying limited recovery on deformed membranes, while partial recovery is observed on spherical, undeformed Nwk1–428-coated GUVs. Scale bar is 10 µm. Associated with Movies S1–S3. (D) Quantification of recovery of protein fluorescence for Nwk1–428 and Nwk1–731. The data are a mean of at least 10 independent experiments and the error bars indicate ± s.e.m. (E) Quantification of protein fluorescence recovery according to vesicle morphology. One-way ANOVA *p<0.05,**p<.005,***p<.001.