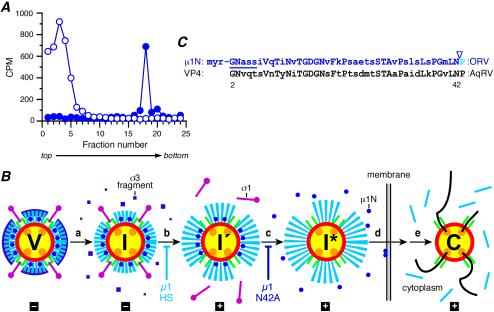
FIG. 7.
μ1N release from ISVP*s, model, and sequences. (A) μ1N release from ISVP*s. [3H]myristate-labeled ISVPs were incubated with either 300 mM NaCl (solid circles) or 300 mM CsCl (open circles) in the presence of 0.5% Triton X-100 for 30 min at 32°C. The particles were then purified on a 1.25- to 1.45-g/cm3 CsCl density gradient. The gradients were fractionated, and the radioactivity (in counts per minute) in each sample was measured by scintillation counting. The results of a representative experiment are shown. (B) Updated model of reovirus uncoating in vitro and during cell entry. Interactions of viral particles and proteins with cell surface receptors and possible localizations to specific subcellular compartments are not included in the diagram. The proposed uncoating intermediates and fates of the outer capsid proteins—μ1N (blue), μ1C (cyan), σ1 (magenta), and σ3 (purple)—are shown. (Step a) The virion (V) undergoes proteolytic processing to generate the ISVP (I). In ISVPs, the σ3 protein has been degraded, and its differently sized fragments have been released. (Step b) The ISVP then undergoes a major structural transition to the ISVP′ (I′). ISVP′s lack σ1 and contain an altered, hydrophobic conformer of μ1. Particles containing μ1HS are blocked at step b (18). (Step c) The ISVP′ then undergoes a further transition to the ISVP* (I*), during which the remaining μ1N/μ1C cleavage occurs and the μ1N peptide is released. As suggested in the diagram (by the increase in particle diameter), further conformational changes in μ1C seem likely to accompany this transition. Particles containing μ1N42A are blocked at step c. (Steps d and e) The released μ1N peptides, putatively in concert with other portions of μ1C remaining in the ISVP*, effects membrane penetration (step d), and transcriptionally active core particles (C) are ultimately released into the cytoplasm (step e). The capacity of each particle type to mediate viral transcription is indicated below the diagram (+, able to mediate; −, unable to mediate). Note that cleavage of μ1C at the δ-φ junction during generation of ISVPs is not shown in this diagram, because the fate of φ during and after membrane penetration remains unknown. The cyan lines accompanying the core after step e specifically represent the released δ fragment (18). (C) Sequence comparison of the N-terminal regions of orthoreovirus (ORV) μ1 and aquareovirus (AqRV) VP4 proteins. Amino acids 2 to 43 of μ1 are shown on top, with corresponding residues of VP4 aligned below. Identical residues are shown in uppercase. The myristoylation consensus sequence in both proteins is indicated by the line. The N-terminal N-myristoyl group of μ1 is labeled (myr), and the site of the putative autocleavage is indicated by the arrowhead. The myristoyl group plus amino acids 2 to 42 constitute the μ1N peptide.