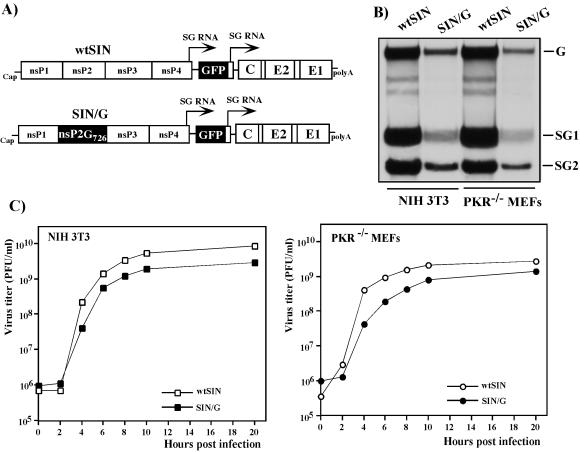
FIG. 1.
Schematic representation of double subgenomic viral genomes and analysis of viral RNA replication and virus growth in NIH 3T3 cells and PKR−/− MEFs. (A) In both genomes, the first SP was driving transcription of GFP-encoding RNA and the second was driving the expression of natural 26S subgenomic RNA encoding SIN structural genes derived from the SIN TE12 strain. The SIN/G variant differed from wtSIN by one amino acid in nsP2, P726 → G. (B) NIH 3T3 cells and PKR−/− MEFs in six-well Costar plates were infected with wtSIN and SIN/G variants at an MOI of 20 PFU/cell. At 5 h postinfection, the media were replaced by 0.8 ml of alpha MEM supplemented with 10% FBS, dactinomycin (1 μg/ml), and [3H]uridine (20 μCi/ml). After 3 h of incubation at 37°C, RNAs were isolated and analyzed by agarose gel electrophoresis as described in Materials and Methods. G, SG1, and SG2 indicate the positions of genomic RNA and subgenomic RNAs 1 and 2, respectively. (C) NIH 3T3 cells and PKR−/− MEFs in six-well Costar plates were infected with wtSIN and SIN/G variants at an MOI of 20 PFU/cell. At the indicated times, the media were replaced and virus titers were determined as described in Materials and Methods.