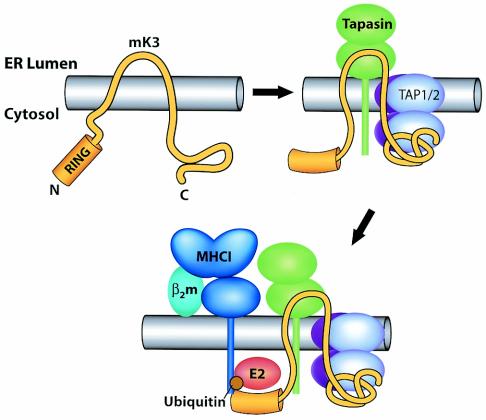
FIG. 8.
Proposed proximity model depicting how mK3 interacts with host proteins to specifically target the rapid degradation of class I proteins. Upper left, nonconformed mK3. Upper right, mK3 forms a complex with TAP/tapasin in a class I-independent manner. The association of mK3 with TAP/tapasin is primarily mediated through the C-terminal domain of mK3, and this interaction may induce a conformational change within mK3 that not only stabilizes mK3 but also restricts the mobility of its RING-CH domain in the N terminus to achieve the proximity, conformation, and/or orientation required for its E3 ligase function. In this model, tapasin, TAP1, and TAP2 are all essential components of this functional interaction with mK3, although our results suggest that TAP1 and -2 are the direct binding partners of mK3. Bottom, an ER-related E2-conjugating enzyme and/or another necessary factor(s) are recruited by the interactions with the N-terminal region of mK3 (both within and outside the RING-CH domain). These interactions result in the conjugation of Ub to the class I molecules. The Ub conjugation is specific for class I proteins due to a required proximity and/or conformation or orientation imposed by mK3 binding to tapasin/TAP1-tapasin/TAP2 complexes. Thus, substrate specificity is determined by interaction with TAP/tapasin, rather than a specific targeting sequence in the tail of class I.