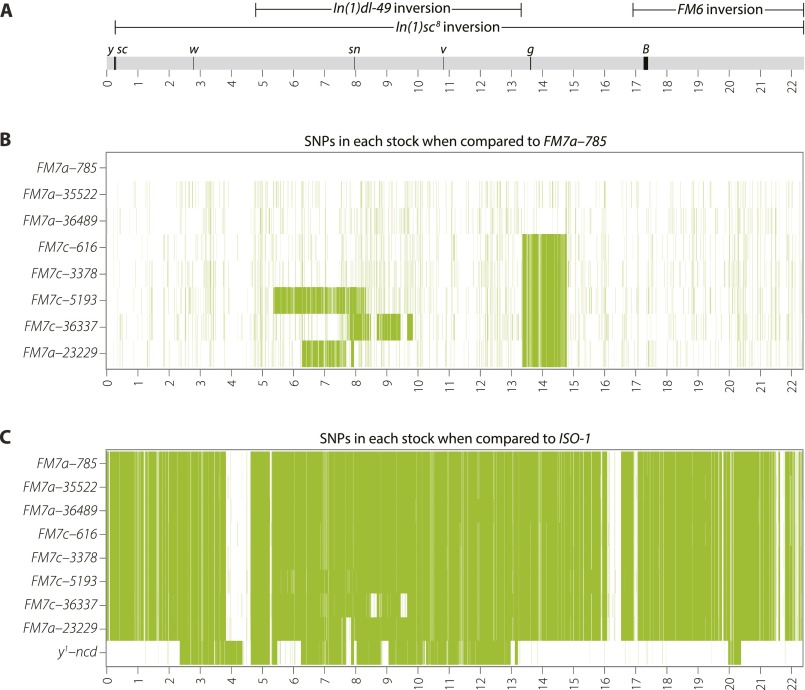
Fig. S1.
Polymorphisms are evident both among FM7 stocks and relative to the ISO-1 reference genome. (A) Schematic of the WT X chromosome, showing the locations of inversions (oriented with respect to the reference genome, not FM7), marker genes, and Release 5 genome coordinates (in Mb). (B) Heatmap of SNPs detected in the eight FM7 stocks used in this study when using FM7a-785 as a genome reference. Increased SNP density covering the sn region in stocks FM7c-5193, FM7c-36337, and FM7a-23229 indicates the region replaced by a DCO event. Note the increased SNP density between 13,369,185 Mb and 14,812,237 Mb present in all FM7c stocks (and the mislabeled FM7a-23229) that defines the haplotype containing g4 present on FM7c. (C) Heatmap of SNPs detected among all FM7 stocks and y1-ncdD compared with the ISO-1 reference genome. Sequence diversity among the eight FM7 stocks is apparent at this scale as differing levels of SNP density surrounding the sn locus. Blocks of similarity between all FM7’s and ISO-1 suggest a common ancestor for these regions. Blocks of diminished SNP density (in white) due to shared ancestry between FM7a-23229 and y1-ncdD are shown in Fig. 3D as an apparent absence of SNPs.