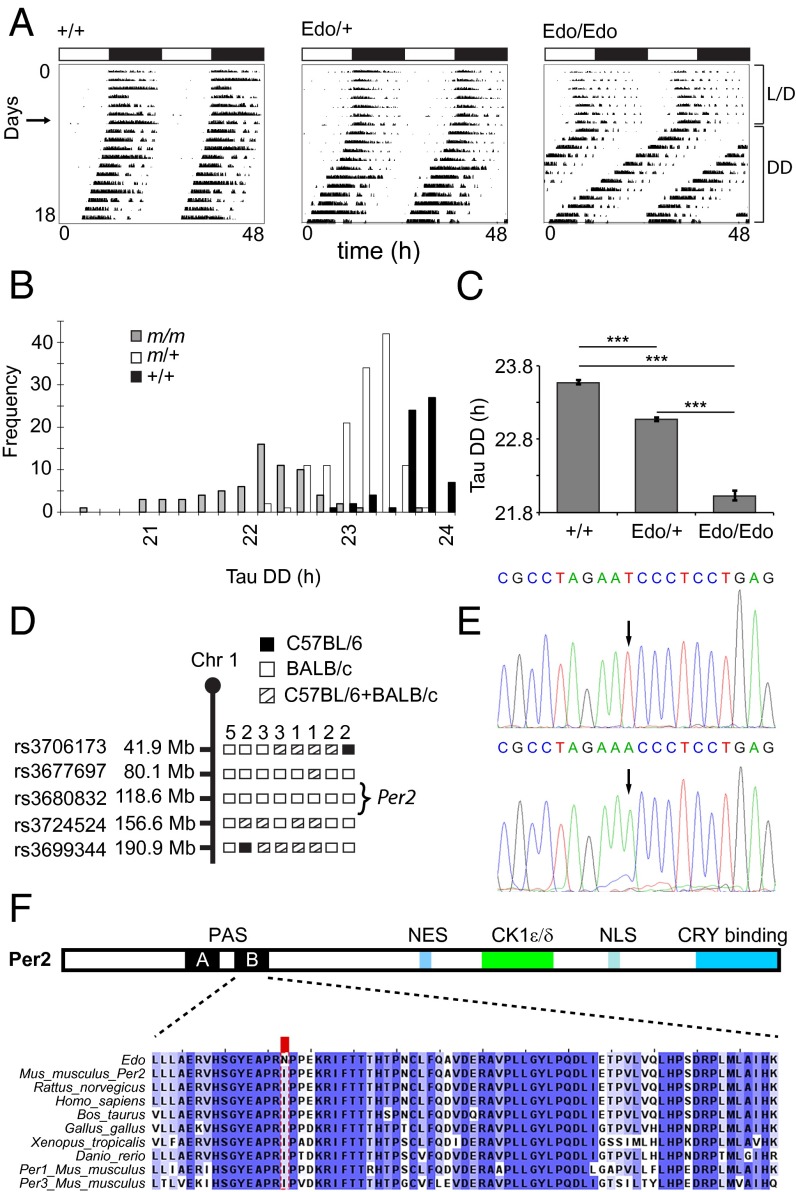
Fig. 1.
Phenotype and cloning of the Per2Edo mutation. (A) Representative double-plotted actograms of wheel-running activity of +/+, Edo/+, and Edo/Edo mice, recorded under light/dark (L/D) conditions (days 1–7) followed by constant darkness (DD). Horizontal bars at the top indicate L/D conditions over the first 7 d. (B) Frequency distribution of the endogenous period [Tau DD (τDD)] of WT (+/+, black), Edo/+ (m/+, white), and Edo/Edo (m/m, gray) littermates. (C) Free-running period of mutants and littermate controls in constant darkness (mean ± SEM; n = 66, 134, 70; one-way ANOVA, P < 0.001; ***Bonferroni pairwise comparisons, P < 0.001). (D) Graphical representation of chromosome 1 (Chr 1) showing positions of SNP markers used to genotype the affected animals. Informative haplotypes for phenotypically homozygous animals are indicated. (E) Chromatograms from resequencing the Per2 gene in +/+ (Top) and Edo/Edo (Bottom) DNA. (F) Functional domains of PER2 (Top) and sequence conservation in PER2 (Bottom) proteins. CK1ε/δ, CK1ε/δ binding domain; NES, nuclear export signal; NLS, nuclear localization signal. The protein sequence alignment of the interdomain linker and PAS-B domain showing the I324N substitution (red box) in Per2Edo is illustrated. High sequence conservation of the interdomain linker and PAS-B domain is evident in vertebrate proteins and in paralogous mouse proteins (PER1, PER3).